Accelerating Bioprocesses: The DBTL Cycle in Modern Metabolic Engineering
This article provides a comprehensive overview of the Design-Build-Test-Learn (DBTL) cycle, a foundational framework in metabolic engineering for developing microbial cell factories.
Accelerating Bioprocesses: The DBTL Cycle in Modern Metabolic Engineering
Abstract
This article provides a comprehensive overview of the Design-Build-Test-Learn (DBTL) cycle, a foundational framework in metabolic engineering for developing microbial cell factories. Tailored for researchers and drug development professionals, it explores the cycle's core principles, from its application in optimizing pathways for compounds like dopamine and fine chemicals to advanced integration with machine learning and automation. The scope covers practical methodologies, common troubleshooting strategies, and comparative analyses of emerging paradigms, offering a holistic guide for implementing efficient, iterative strain engineering to advance sustainable biomanufacturing and therapeutic development.
The DBTL Framework: Core Principles and Evolutionary Impact in Metabolic Engineering
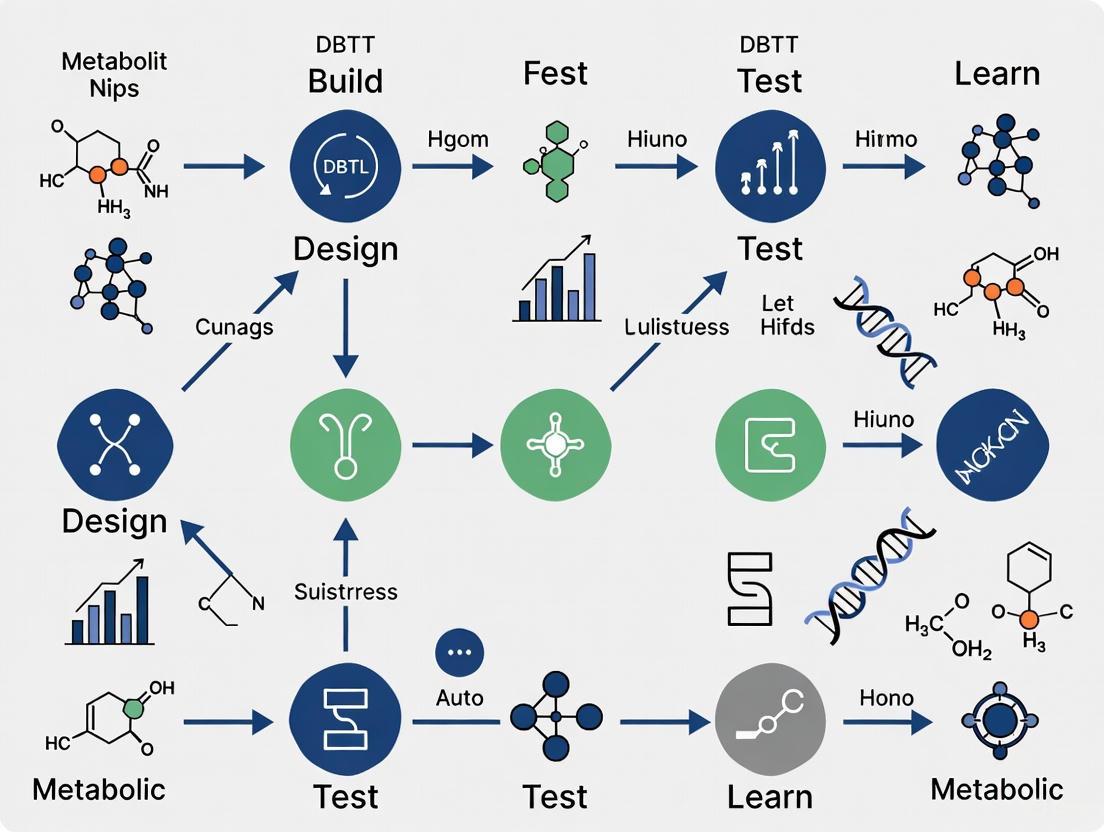
The Design-Build-Test-Learn (DBTL) cycle is a systematic, iterative framework that has become a cornerstone of modern metabolic engineering and synthetic biology. It provides a structured approach to optimizing microbial cell factories for the production of valuable compounds, moving beyond traditional trial-and-error methods. By cycling through these four phases, researchers can progressively refine genetic designs, incorporate knowledge from previous iterations, and accelerate the development of economically viable bioprocesses [1] [2]. This guide details the core principles and technical execution of each phase within the context of metabolic engineering research.
The Design Phase
The Design phase involves the strategic planning of genetic modifications to achieve a specific metabolic engineering objective, such as increasing the yield of a target molecule.
Core Objectives and Methodologies
The primary goal is to define which genetic parts to use and how to assemble them to optimize metabolic flux. This includes selecting pathways, enzymes, and regulatory elements.
- Pathway Identification and Selection: Researchers use computational tools to identify biosynthetic gene clusters or perform retrosynthesis to find potential metabolic pathways for the product of interest [2].
- Component Selection: This involves choosing specific enzymes (e.g., codon-optimized coding sequences), promoters, ribosomal binding sites (RBS), and terminators. Libraries of characterized parts are often used to create combinatorial diversity [1] [2].
- Host Strain Engineering: Genome-scale models can identify beneficial host modifications, such as knocking out competing pathways or derepressing feedback inhibition to increase precursor supply [2] [3].
Experimental Protocol: Informing Design with In Vitro Testing
A "knowledge-driven" DBTL cycle uses upstream in vitro experiments to guide the initial design, saving resources.
- Objective: Pre-validate the functionality of a metabolic pathway and assess different enzyme expression levels before committing to in vivo strain construction [3].
- Method:
- Clone the genes of the target pathway (e.g.,
hpaBCandddcfor dopamine production) into plasmids suitable for cell-free protein synthesis (CFPS). - Express the pathway enzymes in a crude cell lysate system derived from a suitable production host (e.g., E. coli).
- Supplement the reaction buffer with necessary precursors (e.g., L-tyrosine) and cofactors.
- Measure the production of the target metabolite (e.g., L-DOPA or dopamine) to determine the most effective enzyme ratios [3].
- Clone the genes of the target pathway (e.g.,
- Outcome: The results from the in vitro test provide a mechanistic understanding and inform the selection of RBS libraries or promoters for the subsequent in vivo Build phase [3].
Visualization: Knowledge-Driven DBTL Workflow
The following diagram illustrates the integrated workflow where the Design phase is informed by preliminary in vitro experimentation.
The Build Phase
The Build phase is the physical construction of the genetically engineered organism as specified in the Design phase.
Core Objectives and Methodologies
This phase focuses on the high-throughput assembly of DNA constructs and their introduction into the microbial chassis.
- DNA Assembly: Techniques include Gibson assembly, Golden Gate assembly, and others to combine multiple DNA parts into a functional plasmid or chromosomal integration [4].
- Strain Engineering: Methods like CRISPR/Cas9, MAGE (multiplex automated genome engineering), and transformation are used to introduce the genetic constructs into the production host [2].
- Library Generation: For combinatorial optimization, researchers build diverse variant libraries by employing RBS engineering, promoter engineering, or site-saturation mutagenesis to create a spectrum of expression levels for pathway enzymes [1] [3].
Research Reagent Solutions
The following table details essential materials and reagents used in the Build phase for metabolic engineering.
Table 1: Key Research Reagents for the Build Phase
| Reagent / Solution | Function in the Build Phase |
|---|---|
| Plasmid Backbones (e.g., pSEVA, pET) | Standardized vectors for gene expression; often contain selection markers (e.g., antibiotic resistance) and origins of replication [4] [3]. |
| Synthesized Gene Fragments | Codon-optimized coding sequences for the enzymes of the heterologous pathway [4] [3]. |
| Restriction Enzymes & Ligases | Molecular tools for cutting and joining DNA fragments in traditional cloning. |
| Assembly Mixes (e.g., Gibson Assembly Mix) | Enzyme mixes for seamless, homology-based assembly of multiple DNA fragments [4]. |
| Competent Cells | Chemically or electro-competent microbial cells (e.g., E. coli DH5α for cloning, production strains like E. coli FUS4.T2) prepared for DNA uptake [3]. |
The Test Phase
The Test phase involves the cultivation of the built strains and the analytical measurement of their performance.
Core Objectives and Methodologies
The goal is to acquire quantitative data on strain performance, including titer, yield, productivity (TYR), and other functional characteristics.
- Cultivation: Strains are cultivated in microplates or bioreactors under defined conditions to evaluate performance in a relevant bioprocess setting [1] [3].
- Analytical Techniques: A hierarchy of methods is used, balancing throughput and informational depth.
- High-Throughput Screening (HTS): Biosensors coupled to fluorescence or colorimetric outputs, or fluorescent-activated cell sorting (FACS), can rapidly assay thousands of variants for target molecule production [2].
- Target Molecule Detection: Chromatographic methods (GC, LC) coupled with mass spectrometry (MS) or UV detection provide confident identification and precise quantification of the target molecule and pathway intermediates, though at a lower throughput [2].
- Omics Analysis: For a limited number of top-performing strains, deep omics analysis (transcriptomics, proteomics, metabolomics) provides a systems-level view to identify pathway bottlenecks and host interactions [2].
Quantitative Data from Testing
The following table summarizes key performance metrics and analytical methods used in the Test phase.
Table 2: Key Performance Metrics and Analytical Methods in the Test Phase
| Performance Metric | Description | Common Analytical Methods |
|---|---|---|
| Titer | Concentration of the target product (e.g., in mg/L or g/L) [3]. | HPLC, LC-MS, GC-MS [2]. |
| Yield | Amount of product formed per amount of substrate consumed (e.g., mg/gbiomass) [3]. | HPLC, LC-MS combined with biomass measurement [2]. |
| Productivity | Titer achieved per unit of time (e.g., mg/L/h). | Calculated from titer and fermentation time. |
| Specificity/Sensitivity | Key for biosensors; measures uniqueness and detection limit for a target molecule [4]. | Plate reader assays (fluorescence, luminescence) [4] [2]. |
The Learn Phase
The Learn phase is the analytical core of the cycle, where data from the Test phase is interpreted to generate actionable knowledge for the next Design phase.
Core Objectives and Methodologies
This phase transforms raw data into predictive models or design rules to propose more effective strains in the next iteration.
- Data Integration and Analysis: Statistical analysis is performed to identify correlations between genetic designs and performance outcomes [3].
- Machine Learning (ML): Supervised learning models (e.g., random forest, gradient boosting) are trained on the combinatorial strain data to predict the performance of untested genetic designs. These models are particularly powerful in the low-data regime typical of early DBTL cycles [1].
- Bottleneck Identification: Omics data and kinetic models are used to pinpoint specific enzymatic or regulatory steps that limit flux through the pathway [1] [2].
- Recommendation Algorithms: Using model predictions, algorithms can automatically recommend a new set of strains to build, balancing the exploration of the design space with the exploitation of high-performing regions [1].
Visualization: The Role of Machine Learning in the DBTL Cycle
Machine learning integrates into the DBTL cycle by learning from tested strains to predict new, high-performing designs.
Case Study: Optimizing Dopamine Production inE. coli
A 2025 study exemplifies the knowledge-driven DBTL cycle for producing dopamine [3].
- Design & Learn (from in vitro): The pathway from L-tyrosine to dopamine via L-DOPA was first tested in a cell-free crude lysate system. This in vitro experiment confirmed pathway functionality and informed the initial in vivo design.
- Build: A library of production strains was built in an L-tyrosine-overproducing E. coli host using RBS engineering to fine-tune the expression levels of the two key enzymes, HpaBC and Ddc.
- Test: The strains were cultivated, and dopamine production was quantified, revealing that the GC content of the Shine-Dalgarno sequence significantly influenced RBS strength and final titer.
- Learn & Re-Design: Data from the first in vivo cycle was analyzed, leading to the development of a high-performance strain.
- Outcome: The optimized strain achieved a dopamine titer of 69.03 ± 1.2 mg/L, a 2.6-fold improvement over the state-of-the-art, demonstrating the power of the iterative DBTL framework [3].
The Design-Build-Test-Learn cycle is a powerful, iterative engine for metabolic engineering. Its effectiveness is heightened by the integration of upstream knowledge, advanced analytics, and machine learning. As the field evolves, new paradigms like LDBT—where machine learning pre-trained on large datasets precedes design—and the use of rapid cell-free systems for building and testing promise to further accelerate the engineering of biological systems [5].
The Role of DBTL in Systems Metabolic Engineering
The Design-Build-Test-Learn (DBTL) cycle represents a core engineering framework in synthetic biology and systems metabolic engineering, enabling the systematic and iterative development of microbial cell factories. This rational approach has revolutionized our ability to reprogram organisms for sustainable production of valuable compounds, from pharmaceuticals to fine chemicals. Systems metabolic engineering integrates tools from synthetic biology, enzyme engineering, omics technologies, and evolutionary engineering within the DBTL framework to optimize metabolic pathways with unprecedented precision [6]. The power of the DBTL cycle lies in its iterative nature—each cycle generates data and insights that inform subsequent designs, progressively optimizing strain performance while simultaneously expanding biological understanding.
As synthetic biology has matured over the past two decades, the DBTL cycle has become increasingly central to biological engineering pipelines. Technical advancements in DNA sequencing and synthesis have dramatically reduced costs and turnaround times, removing previous barriers in the "Design" and "Build" stages [7]. Meanwhile, the emergence of biofoundries with automated high-throughput systems has transformed "Testing" capabilities, though the "Learn" phase has presented persistent challenges due to the complexity of biological systems [7]. Recent integration of machine learning (ML) and artificial intelligence (AI) promises to finally overcome this bottleneck, potentially unleashing the full potential of predictive biological design [8] [7] [9]. This technical guide examines the current state of DBTL implementation in systems metabolic engineering, providing researchers with practical methodologies and insights for advancing microbial strain development.
The Four Phases of the DBTL Cycle
Design Phase
The Design phase initiates the DBTL cycle, encompassing computational planning and in silico design of biological systems. This stage has been revolutionized by sophisticated software tools that enable precise design of proteins, genetic elements, and metabolic pathways. For any target compound, tools like RetroPath and Selenzyme facilitate automated pathway and enzyme selection by analyzing known biochemical routes and evaluating enzyme candidates based on sequence similarity, phylogenetic analysis, and known biochemical characteristics [10]. The design process involves multiple integrated components: Protein Design (selecting natural enzymes or designing novel proteins), Genetic Design (translating amino acid sequences into coding sequences, designing ribosome binding sites, and planning operon architecture), and Assembly Design (planning plasmid construction with consideration of restriction enzyme sites, overhang sequences, and GC content) [9].
A critical advancement in the Design phase is the application of design of experiments (DoE) methodologies to efficiently explore the combinatorial design space. Researchers can design libraries covering numerous variables—including vector backbones with different copy numbers, promoter strengths, and gene order permutations—then statistically reduce thousands of possible combinations to tractable numbers of representative constructs [10]. For instance, in one documented flavonoid production project, researchers reduced 2592 possible configurations to just 16 representative constructs using orthogonal arrays combined with a Latin square for gene positional arrangement, achieving a compression ratio of 162:1 [10]. This approach allows comprehensive exploration of design parameters without requiring impractical numbers of physical constructs.
Table 1: Key Software Tools for the DBTL Design Phase
| Tool Name | Primary Function | Application in Metabolic Engineering |
|---|---|---|
| RetroPath | Automated pathway selection | Identifies biochemical routes for target compounds |
| Selenzyme | Enzyme selection | Recommends optimal enzymes for pathway steps |
| PartsGenie | DNA part design | Designs ribosome binding sites and coding sequences |
| UTR Designer | RBS engineering | Modulates ribosome binding site sequences |
| TeselaGen | DNA assembly protocol generation | Automates design of cloning strategies |
Build Phase
The Build phase translates in silico designs into physical biological constructs, with modern approaches emphasizing high-throughput, automated DNA assembly and strain construction. Automation plays a crucial role in enhancing precision and efficiency, utilizing automated liquid handlers from platforms such as Tecan, Beckman Coulter, and Hamilton Robotics for high-accuracy pipetting in PCR setup, DNA normalization, and plasmid preparation [9]. DNA assembly methods like ligase cycling reaction (LCR) and Gibson assembly enable seamless construction of complex genetic pathways, with automated worklist generation streamlining the assembly process [10] [8].
Integration with DNA synthesis providers such as Twist Bioscience and IDT (Integrated DNA Technologies) facilitates seamless incorporation of custom DNA sequences into automated workflows [9]. Laboratory Information Management System (LIMS) platforms like TeselaGen's software orchestrate the entire build process, managing protocols and tracking samples across different lab equipment while maintaining robust inventory management systems [9]. The Build phase also encompasses genome editing techniques such as CRISPR-Cas and multiplex automated genome engineering (MAGE), enabling precise chromosomal integration of designed pathways [8]. These automated construction processes significantly reduce human error while increasing throughput—essential factors for exploring complex design spaces in systems metabolic engineering.
Test Phase
The Test phase involves high-throughput characterization of constructed strains to evaluate performance and gather quantitative data. Automated 96-deepwell plate growth and induction protocols enable parallel cultivation of numerous strains under controlled conditions [10]. Analytical chemistry platforms, particularly ultra-performance liquid chromatography coupled to tandem mass spectrometry (UPLC-MS/MS) with high mass resolution, provide sensitive, quantitative detection of target products and key intermediates [10]. Advanced omics technologies further enhance testing capabilities: next-generation sequencing (NGS) platforms like Illumina's NovaSeq enable genotypic verification, while automated mass spectrometry setups such as Thermo Fisher's Orbitrap support proteomic and metabolomic analyses [8] [9].
The Test phase generates extensive datasets requiring sophisticated bioinformatics processing. Custom R scripts and AI-assisted data analysis tools help transform raw analytical data into actionable information [10] [9]. Centralized data management systems act as hubs for collecting information from various analytical and monitoring equipment, integrating test results with design and build data to facilitate comprehensive analysis [9]. This integration is crucial for identifying correlations between genetic designs and phenotypic outcomes, enabling data-driven decisions in subsequent learning phases.
Table 2: Analytical Methods for the DBTL Test Phase
| Method Category | Specific Technologies | Measured Parameters |
|---|---|---|
| Cultivation & Screening | Automated microtiter plate systems, High-throughput bioreactors | Biomass formation, Substrate consumption, General productivity |
| Separations & Mass Spectrometry | UPLC-MS/MS, FIA-HRMS, Orbitrap systems | Target compound titer, Byproduct formation, Intermediate accumulation |
| Sequencing & Genotyping | Next-Generation Sequencing (NGS), Colony PCR | Plasmid sequence verification, Genomic integration validation |
| Multi-omics | RNA-seq, Proteomics, Fluxomics | Pathway activity, Metabolic fluxes, Regulation |
Learn Phase
The Learn phase represents the knowledge-generating component of the cycle, where experimental data is transformed into actionable insights for subsequent design iterations. This phase has traditionally presented the greatest challenges in the DBTL cycle due to biological complexity, but advances in machine learning (ML) and data science are revolutionizing this critical step [7]. Statistical analysis of results identifies the main factors influencing production—for example, in one pinocembrin production study, analysis revealed that vector copy number had the strongest significant effect on production levels, followed by promoter strength for specific pathway enzymes [10].
Machine learning algorithms process vast datasets to uncover complex patterns beyond human detection capacity, enabling accurate genotype-to-phenotype predictions [7] [9]. In one notable application, ML models trained on experimental data guided the optimization of tryptophan metabolism in yeast, demonstrating how computational approaches can accelerate pathway optimization [9]. Explainable ML advances further enhance the learning process by providing both predictions and the underlying reasons for proposed designs, deepening fundamental understanding of biological systems [7]. The learning phase increasingly incorporates mechanistic modeling alongside statistical approaches, particularly through constraint-based metabolic models like flux balance analysis (FBA) and its variants (e.g., pFBA, tFBA) [8]. This combination of data-driven and first-principles approaches creates a powerful framework for extracting maximum knowledge from each DBTL iteration.
Case Studies in Microbial Systems Metabolic Engineering
C5 Platform Chemicals from L-Lysine in Corynebacterium glutamicum
Recent applications of the DBTL cycle to Corynebacterium glutamicum demonstrate its effectiveness in developing microbial cell factories for industrial chemicals. C. glutamicum, traditionally used for amino acid production, has been engineered to produce C5 platform chemicals derived from L-lysine through systematic DBTL iterations [6]. The engineering process began with traditional metabolic engineering to enhance precursor availability, then progressed to advanced systems metabolic engineering integrating synthetic biology tools, enzyme engineering, and omics technologies within the DBTL framework [6].
Researchers applied the DBTL cycle to optimize multiple pathway parameters simultaneously, including enzyme variants, expression levels, and genetic regulatory elements. Through iterative cycling, significant improvements in C5 chemical production were achieved, although specific quantitative metrics were not provided in the available literature [6]. This case exemplifies how the DBTL cycle enables the transformation of traditional industrial microorganisms into sophisticated chemical production platforms through systematic, data-driven engineering.
Flavonoid Production in Escherichia coli
A landmark study published in Communications Biology demonstrated an integrated automated DBTL pipeline for flavonoid production in E. coli, specifically targeting (2S)-pinocembrin [10]. The pathway comprised four enzymes: phenylalanine ammonia-lyase (PAL), 4-coumarate:CoA ligase (4CL), chalcone synthase (CHS), and chalcone isomerase (CHI) converting L-phenylalanine to (2S)-pinocembrin with requirement for malonyl-CoA [10].
Table 3: Progression of Pinocembrin Production Through DBTL Iterations
| DBTL Cycle | Key Design Changes | Resulting Titer (mg/L) | Fold Improvement |
|---|---|---|---|
| Initial Library | 16 representative constructs from 2592 possible combinations | 0.002 - 0.14 | Baseline |
| Second Round | High-copy origin, optimized promoter strengths, fixed gene order | Up to 88 mg/L | 500-fold |
The dramatic improvement resulted from statistical analysis of initial results, which identified vector copy number as the strongest positive factor, followed by CHI promoter strength [10]. Accumulation of the intermediate cinnamic acid indicated PAL enzyme activity was not limiting, allowing strategic focus on other pathway bottlenecks in the second design iteration [10]. This case study exemplifies how the DBTL cycle, particularly when automated, can rapidly converge on optimal designs through data-driven iteration.
Dopamine Production in Escherichia coli
A 2025 study published in Microbial Cell Factories demonstrated a "knowledge-driven DBTL" approach for optimizing dopamine production in E. coli [3]. This innovative methodology incorporated upstream in vitro investigation using cell-free protein synthesis (CFPS) systems to inform initial in vivo strain design, accelerating the overall engineering process. The dopamine biosynthetic pathway comprised two key enzymes: native E. coli 4-hydroxyphenylacetate 3-monooxygenase (HpaBC) converting L-tyrosine to L-DOPA, and L-DOPA decarboxylase (Ddc) from Pseudomonas putida catalyzing the formation of dopamine [3].
The knowledge-driven DBTL approach proceeded through clearly defined experimental stages:
The implementation of this knowledge-driven DBTL cycle resulted in a dopamine production strain achieving 69.03 ± 1.2 mg/L (34.34 ± 0.59 mg/gbiomass), representing a 2.6-fold improvement in titer and 6.6-fold improvement in yield compared to previous state-of-the-art in vivo production systems [3]. Importantly, the approach provided mechanistic insights into the role of GC content in the Shine-Dalgarno sequence on translation efficiency, demonstrating how DBTL cycles can simultaneously achieve both applied and fundamental advances [3].
Essential Research Reagents and Tools for DBTL Implementation
Successful implementation of the DBTL cycle in systems metabolic engineering requires coordinated use of specialized reagents, software, and hardware platforms. The following toolkit details essential resources for establishing DBTL capabilities.
Table 4: The Scientist's Toolkit for DBTL Implementation
| Category | Specific Items | Function & Application |
|---|---|---|
| DNA Assembly & Synthesis | Twist Bioscience, IDT, GenScript oligos | Provides high-quality synthetic DNA fragments for pathway construction |
| Cloning Vectors | pET system, pJNTN, Custom combinatorial vectors | Serves as backbone for pathway expression with tunable copy numbers |
| Automated Liquid Handling | Tecan Freedom EVO, Beckman Coulter Biomek | Enables high-throughput, reproducible PCR setup and DNA assembly |
| Analytical Instruments | UPLC-MS/MS, Illumina NovaSeq, Orbitrap MS | Provides quantitative data on metabolites, proteins, and DNA sequences |
| Strain Engineering Tools | CRISPR-Cas9, MAGE, RBS libraries | Enables precise genomic modifications and expression tuning |
| Software Platforms | TeselaGen, CLC Genomics, RetroPath, Selenzyme | Supports in silico design, data management, and machine learning analysis |
Experimental Protocol: Automated DBTL for Pathway Optimization
This section provides a detailed methodological framework for implementing an automated DBTL cycle, based on established protocols from recent literature [10] [3].
Pathway Design and Library Construction
Pathway Identification: Use RetroPath to identify potential biosynthetic routes to your target compound. Input the target SMILES structure and retrieve possible pathways from biochemical databases [10].
Enzyme Selection: Apply Selenzyme to select optimal enzyme sequences for each pathway step based on sequence similarity, phylogenetic analysis, and known biochemical properties [10].
DNA Part Design: Utilize PartsGenie to design ribosome binding sites with varying strengths and codon-optimized coding sequences suitable for your microbial chassis [10].
Combinatorial Library Design:
- Define design parameters: promoter strengths (strong/weak/none), RBS variants, gene order permutations, and vector backbones with different copy numbers
- Apply Design of Experiments (DoE) with orthogonal arrays to reduce library size while maintaining representative coverage
- For the pinocembrin case, 2592 combinations were reduced to 16 constructs (compression ratio 162:1) [10]
Automated Assembly Protocol Generation: Use software like TeselaGen's platform to generate detailed DNA assembly protocols, specifying cloning method (Gibson, Golden Gate, LCR), fragment preparation, and reaction conditions [9].
High-Throughput Strain Construction
DNA Preparation:
- Order synthetic DNA fragments from commercial providers (Twist Bioscience, IDT)
- Amplify parts via PCR using automated liquid handlers
- Normalize DNA concentrations robotically [10]
Automated Assembly:
- Set up ligase cycling reactions (LCR) or Gibson assemblies using robotic platforms
- Follow automated worklists generated by design software
- Transform into appropriate cloning strain (E. coli DH5α commonly used) [10]
Quality Control:
Screening and Analytics
Cultivation:
- Inoculate verified constructs into 96-deepwell plates containing appropriate medium
- Implement automated growth and induction protocols
- Maintain controlled conditions (temperature, shaking) throughout cultivation [10]
Metabolite Extraction:
- Implement automated quenching and extraction protocols
- Use standardized solvent systems for metabolite recovery
- Include quality control samples and internal standards [10]
Quantitative Analysis:
Data Processing:
- Utilize custom R scripts for data extraction and processing
- Normalize measurements to biomass and internal standards
- Perform quality assessment and remove outliers [10]
Data Analysis and Learning
Statistical Analysis:
- Apply ANOVA to identify significant factors influencing production
- Calculate main effects and interaction effects between design parameters
- For the pinocembrin case, vector copy number showed P value = 2.00 × 10^-8, CHI promoter P value = 1.07 × 10^-7 [10]
Machine Learning Application:
Mechanistic Insight Generation:
- Analyze accumulation of intermediates to identify pathway bottlenecks
- Correlative expression levels with flux measurements
- Formulate testable hypotheses for next DBTL cycle [3]
Emerging Trends in DBTL Implementation
The DBTL cycle continues to evolve with several transformative trends shaping its application in systems metabolic engineering. Machine learning integration is becoming increasingly sophisticated, with explainable AI providing both predictions and underlying reasons for proposed designs [7]. Graph neural networks (GNNs) and physics-informed neural networks (PINNs) represent particularly promising approaches for capturing complex biological relationships [8]. Automation and biofoundries are expanding capabilities, with the Global Biofoundry Alliance establishing standards for high-throughput engineering [7]. The emergence of cloud-based platforms for biological design enables enhanced collaboration and data sharing while providing access to advanced computational resources [9].
The knowledge-driven DBTL approach, exemplified by the dopamine production case, represents a significant advancement over purely statistical design [3]. By incorporating upstream in vitro investigation and mechanistic understanding, this strategy accelerates the learning process and reduces the number of cycles required to achieve optimization targets. Similarly, the integration of cell-free systems for rapid pathway prototyping provides a complementary approach to whole-cell engineering, enabling faster design iteration [3].
The DBTL cycle has established itself as the foundational framework for systems metabolic engineering, enabling systematic optimization of microbial cell factories for sustainable chemical production. Through iterative design, construction, testing, and learning, researchers can progressively refine complex biological systems despite their inherent complexity. The cases reviewed herein—from C5 chemical production in C. glutamicum to flavonoid and dopamine production in E. coli—demonstrate the remarkable effectiveness of this approach across diverse hosts and target compounds.
As DBTL methodologies continue to advance through increased automation, improved computational tools, and enhanced machine learning capabilities, the precision and efficiency of metabolic engineering will further accelerate. These developments promise to unlock new possibilities in sustainable manufacturing, therapeutic development, and fundamental biological understanding, firmly establishing the DBTL cycle as an indispensable paradigm for 21st-century biotechnology.
From Sequential Debottlenecking to Combinatorial Pathway Optimization
The establishment of efficient microbial cell factories for the production of biofuels, pharmaceuticals, and high-value chemicals represents a central goal of industrial biotechnology. Achieving this requires the precise optimization of metabolic pathways to maximize flux toward desired products while maintaining cellular viability. For decades, the predominant framework for this optimization was sequential debottlenecking—a methodical approach where rate-limiting steps in a pathway are identified and alleviated one at a time [11]. This classical approach follows a linear problem-solving logic: identify the greatest constraint, remove it, then identify the next constraint in an iterative manner. While this strategy has yielded successes, it operates under the simplification that pathway bottlenecks act independently, an assumption that often fails in the interconnected complexity of cellular metabolism [12] [13].
The emergence of combinatorial pathway optimization marks a paradigm shift in metabolic engineering, enabled by advances in synthetic biology, DNA synthesis, and high-throughput screening technologies. Rather than addressing constraints sequentially, combinatorial approaches simultaneously vary multiple pathway elements to systematically explore the multidimensional design space of pathway expression and function [12] [13]. This strategy aligns with the modern design-build-test-learn (DBTL) cycle framework, which provides an iterative workflow for strain development. Within this context, combinatorial optimization allows researchers to navigate complex fitness landscapes where interactions between pathway components (epistasis) mean that the optimal combination cannot be found by optimizing individual elements in isolation [14] [1]. The transition from sequential to combinatorial approaches fundamentally transforms how metabolic engineers conceptualize and address pathway optimization, moving from a reductionist to a systems-level perspective.
Sequential Debottlenecking: Principles and Limitations
Core Methodology and Applications
Sequential debottlenecking operates on the principle that any production system contains rate-limiting steps that constrain overall throughput. The methodology follows a two-stage process: first, bottleneck identification, where the specific constraints in a process are pinpointed; and second, bottleneck alleviation, where targeted interventions are made to relieve these constraints [11]. In biomanufacturing, this typically involves analyzing process times and resource utilization across unit operations to identify which steps dictate the overall process velocity. The gold standard for identification involves perturbing cycle times in a discrete event simulation model and observing the impact on key performance indicators such as throughput or cycle time [11].
In practice, sequential optimization of metabolic pathways often involves modulating the expression level of individual enzymes, replacing rate-limiting enzymes with improved variants, or removing competing metabolic reactions. For example, in a simple two-stage production process where an upstream bioreactor step takes 300 hours and a downstream purification takes 72 hours, the bioreactor represents the clear bottleneck. No improvement to the downstream processing can increase overall throughput until the bioreactor cycle time is addressed [11]. This illustrates the fundamental logic of sequential optimization: focus engineering efforts only on the most impactful constraints.
Inherent Limitations in Biological Systems
Despite its logical appeal, sequential debottlenecking faces significant limitations when applied to complex biological systems. A primary challenge is shifting bottlenecks, where alleviating one constraint simply causes another part of the system to become limiting [14]. Metabolic control theory explains this phenomenon: minor improvements in one enzyme often render another enzyme the bottleneck of the pathway [14]. This necessitates multiple iterative cycles of identification and alleviation, making the process time-consuming and potentially costly.
The approach also struggles with epistatic interactions between pathway components, where the effect of modifying one element depends on the state of other elements [14]. In naringenin biosynthesis, for instance, beneficial TAL enzyme mutations identified in a low-copy plasmid context actually decreased production when transferred to a high-copy plasmid [14]. This context-dependence means that improvements identified in isolation may not translate to benefits in the full pathway context. Additionally, sequential methods typically miss global optimum solutions that require coordinated adjustment of multiple parameters simultaneously [1] [13]. Since biological systems exhibit nonlinear behaviors, the sequential approach of holding most variables constant while adjusting one parameter at a time is fundamentally unable to discover synergistic interactions between multiple pathway components.
Table 1: Comparison of Sequential and Combinatorial Optimization Approaches
| Feature | Sequential Debottlenecking | Combinatorial Optimization |
|---|---|---|
| Philosophy | Reductionist, linear | Systems-level, parallel |
| Experimental Throughput | Tests <10 constructs at a time [15] | Tests hundreds to thousands of constructs in parallel [15] |
| Bottleneck Handling | Addresses one bottleneck at a time | Addresses multiple potential bottlenecks simultaneously |
| Optimum Solution | Likely finds local optimum | Capable of finding global optimum [15] |
| Epistasis Accommodation | Poorly accounts for genetic interactions | Explicitly accounts for interactions between components |
| Resource Requirements | Lower per cycle, but more cycles needed | Higher initial investment, potentially fewer cycles |
| Data Efficiency | Generates limited data for modeling | Generates rich datasets for machine learning |
Combinatorial Pathway Optimization: Concepts and Implementation
Theoretical Foundation and Key Advantages
Combinatorial pathway optimization is grounded in the recognition that metabolic pathways constitute complex systems where components interact in non-additive ways. The approach involves the simultaneous variation of multiple pathway parameters to explore a broad design space and identify optimal combinations that would be inaccessible through sequential methods [12] [13]. This strategy explicitly acknowledges that the performance of any individual pathway component depends on its metabolic context, and that the global optimum for pathway function may require specific, coordinated expression levels across multiple enzymes rather than simply maximizing each enzymatic step [1].
The key advantage of combinatorial optimization lies in its ability to overcome epistatic constraints that limit sequential approaches. As demonstrated in the naringenin biosynthesis pathway, beneficial mutations in individual enzymes can exhibit contradictory effects in different genetic contexts, creating a "rugged evolutionary landscape" that traps sequential optimization at local maxima [14]. Combinatorial methods address this challenge by allowing the parallel exploration of multiple enzyme variants and expression levels, effectively smoothing the evolutionary landscape and providing a predictable trajectory for improvement [14]. This capacity makes combinatorial approaches particularly valuable for optimizing nascent pathways where limited a priori knowledge exists about rate-limiting steps or optimal expression balancing.
Implementation Frameworks and Methodologies
Implementing combinatorial optimization requires methodologies for generating genetic diversity and screening resulting variants. The primary strategies for creating combinatorial diversity include: (1) variation of coding sequences through testing homologous enzymes from different organisms or directed evolution; (2) engineering expression levels through promoter engineering, ribosome binding site (RBS) tuning, and gene copy number modulation; and (3) combined approaches that simultaneously address both enzyme identity and expression level [16] [13].
A powerful implementation framework is the biofoundry-assisted strategy for pathway bottlenecking and debottlenecking, which enables parallel evolution of all pathway enzymes along a predictable evolutionary trajectory [14]. This approach uses a "bottlenecking" phase where rate-limiting steps are identified by creating intentional constraints, followed by a "debottlenecking" phase where libraries of enzyme variants are screened under these constrained conditions. This cycle creates selective pressure for improvements that address the specific limitations of the pathway. When combined with machine learning models like ProEnsemble to balance pathway expression, this approach has demonstrated remarkable success, enabling the construction of an E. coli chassis producing 3.65 g/L of naringenin—a significant improvement over previous benchmarks [14].
Diagram 1: Combinatorial Optimization Workflow. This flowchart illustrates the iterative process of intentional bottlenecking, library generation, and machine learning-guided debottlenecking.
The DBTL Cycle: An Integrative Framework for Metabolic Engineering
Components of the DBTL Cycle
The Design-Build-Test-Learn (DBTL) cycle provides a systematic framework for metabolic engineering that integrates both sequential and combinatorial approaches while emphasizing continuous improvement through data-driven learning [1] [3]. In the design phase, researchers specify genetic constructs based on prior knowledge and hypotheses, selecting enzyme variants, regulatory elements, and assembly strategies. The build phase involves physical construction of genetic designs using DNA assembly methods such as Golden Gate assembly, Gibson assembly, or other modular cloning systems [16]. The test phase evaluates constructed strains for performance metrics such as titer, yield, productivity, and growth characteristics. Finally, the learn phase analyzes generated data to extract insights and inform the next design cycle [1] [3].
The power of the DBTL framework lies in its iterative nature and capacity for knowledge accumulation. Each cycle generates data that improves understanding of pathway behavior and constraints, allowing progressively more sophisticated interventions in subsequent cycles. This iterative refinement is particularly powerful when combined with automation and machine learning, enabling the semi-autonomous optimization of complex pathways [1]. The DBTL cycle also provides a structure for integrating different optimization strategies—using combinatorial approaches for broad exploration of the design space initially, then applying more targeted sequential interventions once key constraints are identified.
Knowledge-Driven DBTL and Machine Learning Integration
Recent advances in DBTL implementation emphasize knowledge-driven approaches that maximize learning from each cycle. For example, incorporating upstream in vitro investigations using cell-free protein synthesis systems can provide mechanistic insights before committing to full cellular engineering [3]. This approach was successfully applied in optimizing dopamine production in E. coli, where in vitro testing informed RBS engineering strategies that resulted in a 2.6 to 6.6-fold improvement over previous benchmarks [3].
Machine learning integration represents another major advancement in DBTL implementation. ML algorithms such as gradient boosting and random forest models can analyze complex datasets from combinatorial screens to predict optimal pathway configurations [1]. These models are particularly valuable in the "learn" phase of the DBTL cycle, where they can identify non-intuitive relationships between pathway components and performance. When combined with automated recommendation tools, machine learning enables the semi-autonomous prioritization of designs for subsequent DBTL cycles, dramatically accelerating the optimization process [1]. The simulated DBTL framework allows researchers to test different machine learning methods and experimental strategies in silico before wet-lab implementation, optimizing the allocation of experimental resources [1].
Table 2: Key Experimental Methodologies in Combinatorial Pathway Optimization
| Methodology | Key Features | Applications | Throughput |
|---|---|---|---|
| Golden Gate Assembly | Type IIS restriction enzyme-based; efficient multi-fragment assembly [15] | Pathway construction with standardized parts | Moderate to high |
| RBS Engineering | Modulation of translation initiation rates; precise fine-tuning [3] | Balancing polycistronic operons; metabolic tuning | High |
| CRISPR Screening | Pooled or arrayed screening; genome-scale functional genomics [17] | Identification of fitness-modifying genes; tolerance engineering | Very high |
| Promoter Engineering | Variation of transcriptional initiation rates; library generation [13] | Balancing multi-gene pathways; dynamic regulation | High |
| MAGE (Multiplex Automated Genome Engineering) | In vivo mutagenesis; continuous culturing [14] | Directed evolution of pathway enzymes; genome refinement | High |
| Biosensor-Based Screening | Genetically encoded metabolite sensors; fluorescence-activated sorting [17] [12] | High-throughput screening of production strains | Very high |
Research Toolkit: Essential Reagents and Technologies
The implementation of combinatorial pathway optimization relies on a suite of specialized reagents, tools, and technologies that enable high-throughput genetic manipulation and screening.
Table 3: Essential Research Reagent Solutions for Combinatorial Optimization
| Reagent/Tool | Function | Application Example |
|---|---|---|
| Modular Cloning Toolkits | Standardized genetic parts for combinatorial assembly [16] | Golden Gate-based systems (MoClo, GoldenBraid) |
| CRISPRi/a Screening Libraries | Pooled guide RNA libraries for functional genomics [17] | Identification of gene targets affecting production or tolerance |
| Whole-Cell Biosensors | Transcription factor-based metabolite detection [17] [12] | High-throughput screening of strain libraries via FACS |
| Cell-Free Protein Synthesis Systems | In vitro transcription-translation for rapid prototyping [3] | Pre-testing enzyme combinations before in vivo implementation |
| Orthogonal Regulator Systems | TALEs, zinc fingers, or dCas9-based transcription control [12] | Independent regulation of multiple pathway genes |
| Barcoded Assembly Systems | Tracking library variants via DNA barcodes [12] | Multiplexed analysis of strain library performance |
| 1,2,4-Trimethoxy-5-nitrobenzene | 1,2,4-Trimethoxy-5-nitrobenzene, CAS:14227-14-6, MF:C9H11NO5, MW:213.19 g/mol | Chemical Reagent |
| 4-Nitrodiazoaminobenzene | 4-Nitrodiazoaminobenzene | High-Purity Research Chemical | High-purity 4-Nitrodiazoaminobenzene for research applications. For Research Use Only. Not for human or veterinary use. |
Comparative Analysis: Strategic Implementation Considerations
Choosing between sequential and combinatorial optimization approaches requires careful consideration of project constraints and goals. Sequential debottlenecking may be preferable when resources are limited and high-throughput capabilities are unavailable, when working with well-characterized pathways where major bottlenecks are already known, or when regulatory constraints necessitate minimal genetic modification [11] [15]. The methodical nature of sequential approaches also makes them suitable for educational settings or when establishing foundational protocols.
Combinatorial optimization is particularly advantageous when addressing complex pathways with suspected epistatic interactions, when high-throughput capabilities are available, when optimizing entirely novel pathways with limited prior knowledge, and when pursuing aggressive performance targets that require global optima rather than incremental improvements [12] [13] [15]. The higher initial investment in combinatorial approaches can be offset by reduced overall development time and superior final outcomes.
In practice, many successful metabolic engineering projects employ hybrid strategies that combine elements of both approaches. An initial combinatorial screen might identify promising regions of the design space, followed by more targeted sequential optimization to refine specific pathway components. This hybrid approach leverages the strengths of both methodologies while mitigating their respective limitations.
Diagram 2: Strategy Selection Workflow. This diagram illustrates decision pathways for implementing sequential, combinatorial, or hybrid optimization approaches.
The evolution from sequential debottlenecking to combinatorial pathway optimization represents significant progress in metabolic engineering methodology. While sequential approaches provide a methodical framework for addressing clear rate-limiting steps, combinatorial strategies offer a more powerful means of navigating the complex, interactive nature of metabolic networks. The DBTL cycle serves as an integrative framework that unites these approaches, emphasizing iterative improvement and data-driven learning.
Future advancements in combinatorial optimization will likely focus on enhancing automation, refining machine learning algorithms, and developing more sophisticated high-throughput screening methodologies. As these technologies mature, the distinction between sequential and combinatorial approaches may blur further, giving rise to adaptive optimization strategies that dynamically adjust their approach based on emerging data. Regardless of specific methodology, the fundamental goal remains the efficient development of microbial cell factories that can sustainably produce the chemicals, materials, and therapeutics society needs.
DBTL as a Driver for Sustainable Biomanufacturing
The Design-Build-Test-Learn (DBTL) cycle represents a systematic, iterative framework central to advancing synthetic biology and metabolic engineering. This engineering-based approach has become fundamental for developing microbial cell factories that convert renewable substrates into valuable chemicals, thereby supporting the transition toward a circular bioeconomy. The global market for biomanufacturing is projected to reach $30.3 billion by 2027, underscoring the economic significance of these technologies [18]. Within the context of sustainable biomanufacturing, the DBTL cycle enables researchers to rapidly engineer microorganisms that can replace fossil-based production processes, mitigate anthropogenic greenhouse gas emissions, and utilize waste streams as feedstocks [19] [20]. However, conventional strain development often faces a "valley-of-death" where promising innovations stall due to the overwhelming complexity of potential genetic manipulations and the time-consuming, trial-and-error nature of screening [19]. This challenge has catalyzed the development of next-generation DBTL frameworks that integrate automation, bio-intelligence, and machine learning to dramatically increase the speed and success rate of creating efficient biocatalysts for industrial applications.
The Core DBTL Cycle and Its Evolution
The Fundamental Workflow
The traditional DBTL cycle comprises four distinct but interconnected phases:
- Design: Researchers define objectives for the desired biological function and design genetic parts or systems using computational tools, domain knowledge, and modeling [5]. This phase includes protein design, genetic design (translating amino acid sequences into coding sequences, designing ribosome binding sites, and planning operon architecture), and assay design [9].
- Build: DNA constructs are synthesized and assembled into plasmids or other vectors, then introduced into a characterization system such as bacterial, eukaryotic, or cell-free systems [5]. This phase relies on high-precision laboratory automation including liquid handlers, PCR setup, DNA normalization, and plasmid preparation [9].
- Test: Engineered biological constructs are experimentally measured for performance through quantitative screening, often involving ultra-performance liquid chromatography coupled to tandem mass spectrometry, next-generation sequencing, or automated plate readers [9] [10].
- Learn: Data collected during testing is analyzed and compared to initial objectives to inform the next design round, increasingly using statistical methods and machine learning to identify relationships between design factors and production levels [10] [5].
This workflow closely resembles approaches used in established engineering disciplines such as mechanical engineering, where iteration involves gathering information, processing it, identifying design revisions, and implementing those changes [5].
The Bio-Intelligent DBTL (biDBTL) Framework
Recent advances have evolved the conventional DBTL cycle into a bio-intelligent DBTL (biDBTL) approach that bridges microbiology, molecular biology, biochemical engineering with informatics, automation engineering, and mechanical engineering [19]. This interdisciplinary framework incorporates:
- Novel metrics, biosensors, and bioactuators for bi-directional communication at biological-technical interfaces [20]
- Digital twins mimicking cellular and process levels [20]
- Integration of artificial intelligence to improve prediction quality and enable hybrid learning [20]
- Cellular twining through enzyme-constraint genome-scale metabolic models [19]
The bio-intelligent approach rigorously applied in projects like BIOS aims to accelerate and improve conventional strain and bioprocess engineering, opening the door to decentralized, networked collaboration for strain and process engineering [20].
The Emergence of LDBT: A Paradigm Shift
A more radical transformation proposes reordering the cycle to LDBT (Learn-Design-Build-Test), where machine learning precedes design [5]. This paradigm shift leverages:
- Zero-shot predictions from protein language models (e.g., ESM and ProGen) trained on evolutionary relationships between protein sequences [5]
- Structure-based deep learning tools (e.g., ProteinMPNN) that take entire protein structures as input and predict new sequences that fold into that backbone [5]
- Physics-informed machine learning combining predictive power of statistical models with explanatory strength of physical principles [5]
This approach potentially enables a single cycle to generate functional parts and circuits, moving synthetic biology closer to a Design-Build-Work model that relies on first principles, similar to disciplines like civil engineering [5].
Advanced DBTL Methodologies and Technologies
Machine Learning Integration in DBTL Cycles
Machine learning (ML) has become a transformative force in synthetic biology, addressing critical limitations of traditional DBTL cycles that often lead to involution states - iterative trial-and-error that increases complexity without corresponding gains in productivity [18]. ML techniques effectively capture complex patterns and multi-cellular level relations from data that are difficult to explicitly model analytically [18].
Key ML applications in DBTL include:
- Protein language models (ESM, ProGen) that capture long-range evolutionary dependencies within amino acid sequences [5]
- Structure-based design tools (MutCompute, ProteinMPNN) that associate amino acids with their chemical environment or predict sequences for specific backbones [5]
- Functional prediction models for thermostability (Prethermut, Stability Oracle) and solubility (DeepSol) [5]
- Hybrid approaches integrating evolutionary and biophysical information to enhance predictive power [5]
These data-driven techniques can incorporate features from micro-aspects (enzymes and cells) to scaled process variables (reactor conditions) for titer predictions, overcoming limitations of mechanistic models that struggle with highly nonlinear cellular processes under multilevel regulations [18].
Automation and Digital Infrastructure
Automation plays a crucial role in enhancing precision and efficiency across all DBTL phases, transforming biotech R&D workflows through:
- Advanced software platforms (e.g., TeselaGen) that generate detailed DNA assembly protocols, manage high-throughput workflows, and orchestrate laboratory automation [9]
- Automated liquid handlers (Labcyte, Tecan, Beckman Coulter, Hamilton Robotics) enabling high-precision pipetting for PCR setup, DNA normalization, and plasmid preparation [9]
- High-throughput screening systems including plate readers (EnVision Multilabel Plate Reader, BioTek Synergy HTX) and next-generation sequencing platforms (Illumina NovaSeq, Ion Torrent) [9]
- Integration with DNA synthesis providers (Twist Bioscience, IDT, GenScript) streamlining custom DNA sequence integration into lab workflows [9]
The digital infrastructure supporting automated DBTL cycles requires strategic deployment choices between cloud vs. on-premises solutions, each offering distinct advantages for scalability, collaboration, security, and compliance with regulatory requirements [9].
Cell-Free Systems for Accelerated Building and Testing
Cell-free gene expression platforms have emerged as powerful tools for accelerating the Build and Test phases of DBTL cycles. These systems leverage protein biosynthesis machinery from crude cell lysates or purified components to activate in vitro transcription and translation [5]. Key advantages include:
- Rapid protein production (>1 g/L protein in <4 hours) without time-intensive cloning steps [5]
- Scalability from picoliter to kiloliter scales, enabling high-throughput screening [5]
- Production of toxic products that would be incompatible with live cells [5]
- Facile customization of reaction environments and incorporation of non-canonical amino acids [5]
When combined with liquid handling robots and microfluidics, cell-free systems can screen upwards of 100,000 picoliter-scale reactions, generating the massive datasets required for training robust machine learning models [5].
Case Study: Automated DBTL Pipeline for Flavonoid Production
A comprehensive study demonstrated the power of an integrated, automated DBTL pipeline for optimizing microbial production of fine chemicals, specifically targeting the flavonoid (2S)-pinocembrin in Escherichia coli [10]. This case study exemplifies the quantitative improvements achievable through systematic DBTL implementation.
Experimental Design and Workflow
First DBTL Cycle Design Parameters:
- Pathway enzymes: Phenylalanine ammonia-lyase (PAL), chalcone synthase (CHS), chalcone isomerase (CHI), and 4-coumarate:CoA ligase (4CL) [10]
- Expression regulation: Four vector backbones with varying copy numbers (p15a medium copy, pSC101 low copy) and promoters (Ptrc strong, PlacUV5 weak) [10]
- Combinatorial factors: Strong, weak, or no promoter for each intergenic region; 24 permutations of gene order [10]
- Library compression: 2592 possible configurations reduced to 16 representative constructs using design of experiments (DoE) with orthogonal arrays and Latin square for positional arrangement [10]
Build Phase:
- Automated ligase cycling reaction for pathway assembly on robotics platforms [10]
- Commercial DNA synthesis followed by part preparation via PCR [10]
- High-throughput automated plasmid purification, restriction digest, and capillary electrophoresis for quality control [10]
Test Phase:
- HTP 96-deepwell plate-based growth pipeline [10]
- Automated extraction and quantitative screening via fast UPLC coupled to tandem mass spectrometry with high mass resolution [10]
- Custom R scripts for data extraction and processing [10]
Learn Phase:
- Statistical analysis identifying relationships between design factors and production levels [10]
- Vector copy number showed strongest significant effect on pinocembrin levels (P value = 2.00 × 10â»â¸) [10]
- CHI promoter strength had positive effect (P value = 1.07 × 10â»â·) [10]
- Weaker effects observed for CHS, 4CL, and PAL promoter strengths [10]
- High accumulation of intermediate cinnamic acid suggested PAL activity wasn't limiting [10]
Second DBTL Cycle Optimizations:
- High copy number origin (ColE1) selected for all constructs [10]
- CHI position fixed at pathway beginning to ensure direct promoter placement [10]
- 4CL and CHS allowed to exchange positions with no, low, or high strength promoters [10]
- PAL location fixed at the 3' end of the assembly [10]
Quantitative Results and Performance Metrics
Table 1: Performance Improvement Through Iterative DBTL Cycling for Pinocembrin Production in E. coli
| DBTL Cycle | Library Size | Maximum Titer (mg/L) | Key Identified Factors | Statistical Significance (P-value) |
|---|---|---|---|---|
| Initial Cycle | 16 constructs | 0.14 mg/L | Vector copy number | 2.00 × 10â»â¸ |
| CHI promoter strength | 1.07 × 10â»â· | |||
| CHS promoter strength | 1.01 × 10â»â´ | |||
| Second Cycle | Optimized design | 88 mg/L | High copy number backbone | Not reported |
| Strategic CHI placement | Not reported | |||
| Overall Improvement | — | 500-fold increase | — | — |
The application of two iterative DBTL cycles successfully established a production pathway improved by 500-fold, with competitive titers reaching 88 mg Lâ»Â¹, demonstrating the powerful efficiency gains achievable through automated DBTL methodologies [10].
Research Reagent Solutions for DBTL Workflows
Table 2: Essential Research Reagents and Platforms for Advanced DBTL Implementation
| Category | Specific Tools/Platforms | Function in DBTL Pipeline |
|---|---|---|
| DNA Design Software | RetroPath [10], Selenzyme [10], PartsGenie [10] | Automated pathway and enzyme selection, parts design with optimized RBS and coding regions |
| DNA Assembly & Cloning | Ligase Cycling Reaction (LCR) [10], Gibson Assembly [9], Golden Gate Cloning [9] | High-efficiency assembly of DNA constructs with minimal errors |
| Automated Liquid Handlers | Labcyte, Tecan, Beckman Coulter, Hamilton Robotics [9] | High-precision pipetting for PCR setup, DNA normalization, and plasmid preparation |
| DNA Synthesis Providers | Twist Bioscience, IDT, GenScript [9] | Custom DNA sequence synthesis for integration into automated workflows |
| Analytical Screening | UPLC-MS/MS [10], Illumina NovaSeq [9], Thermo Fisher Orbitrap [9] | Quantitative measurement of target products, genotypic analysis, proteomic profiling |
| Cell-Free Systems | In vitro transcription/translation systems [5] | Rapid protein expression without cloning, high-throughput sequence-to-function mapping |
| Machine Learning Tools | ESM [5], ProGen [5], ProteinMPNN [5] | Zero-shot prediction of protein structure-function relationships, library design |
Industrial Applications and Sustainability Impact
Showcase Applications in Sustainable Manufacturing
The BIOS project demonstrates the industrial relevance of advanced DBTL frameworks by focusing on creating Pseudomonas putida producer strains for high-value chemicals from waste streams [19] [20]. Key showcase applications include:
- Production of (hydroxy) fatty acids and PHA from one-carbon (methanol/formate) and lignin waste streams for polyester synthesis [19]
- Terpene production for pharmaceuticals, fragrances, and biofuel precursors [20]
- Methylacrylate production for biodegradable polymers and materials [20]
- Polyolefines production from renewable resources [20]
These applications target highly attractive products with significant potential for reducing anthropogenic greenhouse footprint, supporting the transition from fossil-based production processes to a circular bioeconomy [20].
Environmental and Economic Benefits
Advanced DBTL approaches contribute substantially to sustainability goals through:
- Waste stream valorization: Converting one-carbon compounds (methanol, formate) and lignin derivatives into valuable chemicals [19]
- Fossil resource displacement: Replacing petroleum-based production with biological manufacturing routes [19] [20]
- Reduced greenhouse gas emissions: Mitigating atmospheric COâ‚‚ levels through circular bioeconomy approaches [20]
- Economic viability acceleration: Overcoming the "valley-of-death" in strain engineering through rapid, predictive design cycles [19]
The implementation of bio-intelligent DBTL cycles ultimately paves the way for decentralized bio-manufacturing through autonomous, self-controlled bioprocesses that can operate efficiently at various scales [20].
The DBTL cycle has evolved from a conceptual framework to a powerful, integrated pipeline driving sustainable biomanufacturing forward. The integration of automation, machine learning, and bio-intelligent systems has transformed metabolic engineering from a trial-and-error discipline to a predictive science capable of addressing urgent sustainability challenges. The demonstrated 500-fold improvement in production titers through iterative DBTL cycling underscores the transformative potential of these approaches [10].
Future advancements will likely focus on fully autonomous DBTL systems where artificial intelligence agents manage the entire cycle from design to learning [5]. The emergence of LDBT paradigms suggests a fundamental shift toward first-principles biological engineering, potentially reducing multiple iterative cycles to single, highly efficient design iterations [5]. As these technologies mature, DBTL-driven biomanufacturing will play an increasingly critical role in establishing a circular bioeconomy, reducing dependence on fossil resources, and mitigating the environmental impact of chemical production across diverse industrial sectors.
From Theory to Bioproduction: Implementing DBTL for Pathway and Strain Optimization
The Design-Build-Test-Learn (DBTL) cycle represents a systematic framework for accelerating biological engineering, enabling the rapid optimization of microbial strains for chemical production. This whitepaper explores the implementation of an automated DBTL pipeline for the prototyping of microbial production platforms, using the enhanced biosynthesis of the flavonoid (2S)-pinocembrin in Escherichia coli as a detailed case study. We delineate how the integration of computational design, automated genetic assembly, high-throughput analytics, and machine learning facilitates the efficient optimization of complex metabolic pathways. The application of this pipeline to pinocembrin production resulted in a 500-fold improvement in titers, achieving levels up to 88 mg Lâ»Â¹ through two iterative cycles, demonstrating a compound-agnostic and automated approach applicable to a wide range of fine chemicals [10] [21]. The methodologies, datasets, and engineered strains presented herein provide a blueprint for the application of automated DBTL cycles in metabolic engineering research and industrial biomanufacturing.
The DBTL cycle is an engineering paradigm that has been successfully adapted from traditional engineering disciplines to synthetic biology and metabolic engineering. Its iterative application is central to the rational development of microbial cell factories. In the context of metabolic engineering for natural product synthesis:
- The Design phase involves the in silico selection of biosynthetic pathways and enzymes, followed by the detailed design of genetic constructs using standardized biological parts.
- The Build phase encompasses the physical, and often automated, assembly of these designed genetic constructs into a microbial chassis.
- The Test phase focuses on cultivating the engineered strains and quantitatively evaluating their performance, typically through high-throughput analytical methods.
- The Learn phase uses statistical analysis and machine learning on the generated data to extract insights, identify bottlenecks, and inform the design of the next cycle [10] [8].
Fully automated biofoundries are now operational, leveraging laboratory robotics and sophisticated software to execute these cycles with unprecedented speed and scale. This automation is crucial for exploring the vast combinatorial space of genetic designs, a task that is intractable with manual methods [10]. This whitepaper examines a specific implementation of such a pipeline, detailing its components and efficacy through the lens of optimizing pinocembrin, a key flavonoid precursor, production in E. coli.
The Pinocembrin Production Pathway
(2S)-Pinocembrin is a flavanone that serves as a key branch-point intermediate for a wide range of pharmacologically active flavonoids, such as chrysin, pinostrobin, and galangin [22]. Its microbial biosynthesis from central carbon metabolites requires the construction of a heterologous pathway in E. coli.
- Pathway Overview: The biosynthetic route from L-phenylalanine to (2S)-pinocembrin involves four enzymatic steps:
- Phenylalanine ammonia-lyase (PAL): Deaminates L-phenylalanine to form cinnamic acid.
- 4-coumarate:CoA ligase (4CL): Activates cinnamic acid to cinnamoyl-CoA using ATP.
- Chalcone synthase (CHS): Condenses one molecule of cinnamoyl-CoA with three molecules of malonyl-CoA to form pinocembrin chalcone.
- Chalcone isomerase (CHI): Isomerizes the chalcone to (2S)-pinocembrin [23] [24] [25].
- Host Metabolism Integration: For a sustainable bioprocess, the pathway must be supported by host metabolism. This involves enhancing the supply of the precursors L-phenylalanine, derived from the shikimate pathway, and malonyl-CoA, a central metabolite typically limiting for flavonoid production [22] [24]. Early strategies required the supplementation of expensive phenylpropanoic precursors, but recent advances have enabled production directly from simple carbon sources like glucose or glycerol [23] [22].
The following diagram illustrates the metabolic pathway for pinocembrin production in the engineered E. coli cell, highlighting the key heterologous enzymes and the supporting host metabolism.
Implementing the Automated DBTL Pipeline
The development of a high-titer pinocembrin-producing strain was achieved through an automated, integrated DBTL pipeline. This section details the specific protocols and methodologies employed at each stage.
Design Phase: In Silico Pathway and Library Design
The Design phase leverages a suite of bioinformatics tools to select and design genetic constructs for pathway expression.
- Software Tools: The pipeline uses RetroPath for automated pathway selection from a target compound and Selenzyme for selecting candidate enzymes based on desired biochemical properties [10]. The PartsGenie software is then used to design reusable DNA parts, optimizing ribosome-binding sites (RBS) and codon-usage for the coding sequences [10].
- Combinatorial Library Design: A combinatorial library of 2,592 possible pathway configurations was designed in silico by varying several genetic parameters:
- Vector Backbone: Four options with different copy numbers (e.g., ColE1 - high, p15a - medium, pSC101 - low) and promoters (Ptrc - strong, PlacUV5 - weak).
- Intergenic Regions: Three options for the region preceding each gene: a strong promoter, a weak promoter, or no promoter.
- Gene Order: All 24 permutations of the four genes (PAL, 4CL, CHS, CHI) were considered [10].
- Design of Experiments (DoE): To make the library experimentally tractable, a statistical reduction using orthogonal arrays and a Latin square for gene positioning was applied. This reduced the library from 2,592 to 16 representative constructs, a compression ratio of 162:1, enabling efficient exploration of the design space without the need for ultra-high-throughput construction and screening [10].
Build Phase: Automated DNA Assembly and Construction
The Build phase translates digital designs into physical DNA constructs using automated laboratory workflows.
- DNA Synthesis and Preparation: Protein-coding sequences are commercially synthesized. DNA parts are then prepared via PCR amplification [10].
- Automated Pathway Assembly: Assembly recipes and robotics worklists are generated by the pipeline's software (PlasmidGenie). Pathway assembly is performed using the ligase cycling reaction (LCR) on robotic platforms. This method is highly efficient for assembling multiple DNA fragments [10].
- Quality Control (QC): After transformation into E. coli, candidate plasmid clones are subjected to high-throughput QC. This involves automated plasmid purification, restriction digest analysis via capillary electrophoresis, and finally, sequence verification to confirm assembly accuracy [10]. All designed parts and plasmid assemblies are deposited in a centralized repository (JBEI-ICE) with unique identifiers for sample tracking [10].
Test Phase: High-Throughput Cultivation and Analytics
The Test phase involves cultivating the library of strains and quantifying pathway performance.
- Cultivation Protocol: Constructs are introduced into the production chassis (e.g., E. coli DH5α or engineered derivatives). Automated protocols for 96-deepwell plate cultivation are used, controlling for growth, induction of gene expression, and feeding [10].
- Analytical Chemistry: The detection and quantification of the target product (pinocembrin) and key intermediates (e.g., cinnamic acid) are critical.
- Sample Preparation: Automated extraction of metabolites from culture samples.
- Quantitative Analysis: Analysis is performed using fast ultra-performance liquid chromatography coupled to tandem mass spectrometry (UPLC-MS/MS) with high mass resolution. This provides high sensitivity and specificity [10].
- Data Processing: Custom, open-source R scripts are employed for automated data extraction and processing, converting raw instrument data into quantified metabolite titers [10].
Learn Phase: Data Analysis and Machine Learning
The Learn phase closes the loop by extracting actionable knowledge from the experimental data.
- Statistical Analysis: The measured pinocembrin titers from the 16 constructs are analyzed to identify the main factors influencing production. Techniques like Analysis of Variance (ANOVA) are used to determine the statistical significance of factors like vector copy number, promoter strength for each gene, and gene order [10].
- Insight Generation: In the initial cycle, analysis revealed that vector copy number had the strongest positive effect on pinocembrin titer, followed by the promoter strength for the CHI gene. Interestingly, high levels of the intermediate cinnamic acid accumulated across all constructs, suggesting that PAL activity was not a bottleneck, but that downstream steps might be constrained [10]. These insights directly informed the constraints and focus of the second DBTL cycle.
Case Study Data: Pinocembrin DBTL Cycle Outcomes
The iterative application of the DBTL pipeline led to significant improvements in pinocembrin production. The table below summarizes the quantitative outcomes and key design changes across two reported DBTL cycles.
Table 1: Progression of Pinocembrin Production Through DBTL Iterations
| DBTL Cycle | Key Design Changes & Rationale | Maximum Pinocembrin Titer (mg/L) | Fold Improvement | Key Learning Outcomes |
|---|---|---|---|---|
| Cycle 1 | Initial combinatorial library of 16 constructs (from 2,592 designs) exploring copy number, promoter strength, and gene order. | 0.14 [10] | Baseline | Copy number and CHI promoter strength are most significant. Cinnamic acid accumulates, suggesting downstream bottlenecks. Gene order effect is negligible. |
| Cycle 2 | Library focused on high-copy backbone, fixed CHI position, and varied promoters for 4CL and CHS based on Cycle 1 learnings. | 88 [10] | ~500x | Confirmed the critical importance of high gene dosage and strong expression of CHI and other downstream enzymes. |
Subsequent research, leveraging insights from such DBTL cycles, has further advanced production capabilities by integrating host strain engineering. The following table compares production levels from different metabolic engineering strategies, highlighting the role of the optimized chassis.
Table 2: Advanced Pinocembrin Production through Host Strain Engineering
| Engineering Strategy | Key Host Modifications | Precursor Supplementation? | Maximum Pinocembrin Titer (mg/L) | Citation |
|---|---|---|---|---|
| Modular Pathway Balancing | Overexpression of feedback-insensitive DAHP synthase, PAL, 4CL, CHS, CHI on multiple plasmids. | No (from glucose) | 40.02 [23] [24] | |
| Cinnamic Acid Flux Control | Screening PAL/4CL enzyme combinations; CHS mutagenesis (S165M); malonyl-CoA engineering. | Yes (L-phenylalanine) | 67.81 [25] | |
| Enhanced Chassis Development | Deletion of pta-ackA, adhE; overexpression of CgACC; deletion of fabF; integration of feedback-insensitive ppsA, aroF, pheA. | No (from glycerol) | 353 [22] |
Essential Research Reagents and Solutions
The experimental workflows described rely on a suite of specialized reagents, biological parts, and software tools. The following table catalogues key resources essential for replicating or building upon this automated pipeline prototyping effort.
Table 3: Research Reagent Solutions for DBTL Pipeline Implementation
| Category | Item | Specific Example / Part Number | Function / Application |
|---|---|---|---|
| Enzymes / Genes | Phenylalanine Ammonia-Lyase (PAL) | Arabidopsis thaliana [10], Rhodotorula mucilaginosa [25] | Converts L-phenylalanine to cinnamic acid. |
| 4-Coumarate:CoA Ligase (4CL) | Streptomyces coelicolor [10], Petroselinum crispum [25] | Activates cinnamic acid to cinnamoyl-CoA. | |
| Chalcone Synthase (CHS) | Arabidopsis thaliana [10], Camellia sinensis [22] | Condenses cinnamoyl-CoA with malonyl-CoA. | |
| Chalcone Isomerase (CHI) | Arabidopsis thaliana [10], Medicago sativa [25] | Isomerizes chalcone to (2S)-pinocembrin. | |
| Software Tools | Pathway Design | RetroPath [10] | In silico design of biosynthetic pathways. |
| Enzyme Selection | Selenzyme [10] | Selection of candidate enzymes for pathway steps. | |
| DNA Part Design | PartsGenie [10] | Design and optimization of genetic parts (RBS, coding sequences). | |
| Strain / Chassis | Base Production Chassis | E. coli MG1655 [22], E. coli BL21(DE3) [25] | Common microbial hosts for heterologous expression. |
| Engineered Chassis | SBC010792 (MG1655 derivative with enhanced L-phenylalanine synthesis) [22] | Pre-engineered host with improved precursor supply. | |
| Lab Automation | DNA Assembly | Ligase Cycling Reaction (LCR) [10] | Robust assembly of multiple DNA fragments. |
| Analytics | UPLC-MS/MS [10] | High-throughput, quantitative analysis of metabolites. |
The case study of pinocembrin production in E. coli effectively demonstrates the transformative power of an automated DBTL pipeline for metabolic engineering. The integration of sophisticated computational design, automated robotic workflows, and data-driven learning enabled a rapid 500-fold improvement in production titer within just two cycles. This approach successfully moves beyond the traditional, linear model of strain engineering to a high-dimensional, iterative process that efficiently navigates complex biological design spaces.
Future developments in this field will be driven by deeper integration of machine learning (ML) and artificial intelligence (AI) models that can predict optimal genetic designs from increasingly large and complex datasets [8]. Furthermore, the expansion of biofoundries and the standardization of biological parts and protocols will enhance the transferability and scalability of these pipelines. As these technologies mature, the application of automated DBTL cycles will become the standard for developing microbial cell factories, not only for flavonoids but for a broad spectrum of fine chemicals, therapeutic natural products, and sustainable biomaterials, profoundly impacting drug development and green manufacturing.
The Design-Build-Test-Learn (DBTL) cycle represents a foundational framework in modern metabolic engineering, enabling the systematic development of microbial cell factories for sustainable bioproduction [6]. This iterative process encompasses the rational design of genetic modifications, construction of engineered strains, testing of strain performance, and learning from data to inform the next design cycle. While effective, traditional DBTL approaches often face challenges in initial design efficiency, as the first cycle typically begins without prior mechanistic knowledge, potentially leading to multiple resource-intensive iterations [26].
The emergence of knowledge-driven DBTL workflows addresses this limitation by incorporating upstream mechanistic investigations before embarking on full cycling. This approach leverages in vitro testing systems to generate crucial pathway performance data, providing a rational foundation for initial strain design decisions. This technical guide examines the implementation of this advanced workflow through a case study on dopamine production in Escherichia coli, demonstrating how upstream in vitro investigation accelerates the development of efficient microbial production strains while providing valuable mechanistic insights [26] [27].
Dopamine Biosynthesis Pathway and Engineering Strategy
Pathway Architecture and Key Enzymes
Dopamine biosynthesis in engineered E. coli follows a two-step pathway beginning with the endogenous precursor L-tyrosine:
- Hydroxylation: L-tyrosine is converted to L-3,4-dihydroxyphenylalanine (L-DOPA) by the native E. coli enzyme 4-hydroxyphenylacetate 3-monooxygenase (HpaBC) [26].
- Decarboxylation: L-DOPA is converted to dopamine by the heterologous enzyme L-DOPA decarboxylase (Ddc) from Pseudomonas putida [26].
The host strain requires extensive genomic engineering to ensure adequate precursor supply. Critical modifications include depletion of the transcriptional dual regulator TyrR and mutation of the feedback inhibition in chorismate mutase/prephenate dehydrogenase (TyrA) to increase intracellular L-tyrosine concentrations [26].
Figure 1: Engineered dopamine biosynthesis pathway in E. coli showing key enzymatic steps and precursor engineering targets.
Applications and Production Significance
Dopamine has valuable applications across multiple fields. In emergency medicine, it regulates blood pressure, renal function, and addresses neurobehavioral disorders [26]. Under alkaline conditions, it self-polymerizes into polydopamine, which has applications in cancer diagnosis and treatment, agriculture for plant protection, wastewater treatment for removing heavy metal ions and organic contaminants, and production of lithium anodes in fuel cells as a strong ion and electron conductor [26]. Current industrial-scale dopamine production relies on chemical synthesis or enzymatic systems, both environmentally harmful and resource-intensive processes that microbial production aims to replace [26].
Knowledge-Driven DBTL Workflow Implementation
Integrated In Vitro/In Vivo Approach
The knowledge-driven DBTL cycle incorporates upstream in vitro investigation before embarking on traditional cycling, creating a more efficient and mechanistic strain optimization process [26]. This approach begins with testing enzyme expression levels and pathway functionality in crude cell lysate systems, which provide essential metabolites and energy equivalents while bypassing whole-cell constraints such as membranes and internal regulation [26]. The results from these in vitro studies are then translated to the in vivo environment through high-throughput ribosome binding site (RBS) engineering [26].
Figure 2: Knowledge-driven DBTL workflow integrating upstream in vitro studies with automated cycling.
Experimental Protocols and Methodologies
Crude Cell Lysate System Preparation
The in vitro investigation phase utilizes crude cell lysate systems derived from E. coli production strains [26]:
- Reaction Buffer Composition: 50 mM phosphate buffer (pH 7) supplemented with 0.2 mM FeCl₂, 50 µM vitamin B₆, and 1 mM L-tyrosine or 5 mM L-DOPA as substrates [26].
- System Advantages: Crude cell lysates maintain natural metabolic context with essential cofactors, energy regeneration systems, and metabolite pools that support functional enzyme activity assessment without cellular compartmentalization barriers [26].
- Pathway Validation: Enzyme combinations are tested at varying expression ratios to determine optimal stoichiometry for maximal dopamine flux before genetic implementation [26].
High-Throughput RBS Engineering
Following in vitro optimization, results are translated to in vivo strains through RBS engineering:
- Strategy Focus: Modulation of the Shine-Dalgarno (SD) sequence without interfering with secondary structures to precisely control translation initiation rates (TIR) [26].
- Key Finding: GC content in the SD sequence significantly impacts RBS strength and consequently enzyme expression levels [26].
- Automation: Library construction and screening implemented through biofoundry approaches enabling high-throughput testing of multiple RBS variants [26].
Cultivation Conditions for Production Strains
Optimized dopamine production strains are evaluated under controlled fermentation conditions [26]:
- Base Medium: Minimal medium containing 20 g/L glucose, 10% 2xTY medium, phosphate buffer, MOPS, and essential salts [26].
- Supplementation: 50 µM vitamin B₆, 5 mM phenylalanine, 0.2 mM FeCl₂, and trace element solution [26].
- Culture Conditions: Appropriate antibiotics and inducers (1 mM IPTG) with cultivation at suitable temperature and aeration [26].
Quantitative Results and Performance Metrics
Dopamine Production Performance
Table 1: Comparative dopamine production performance of engineered E. coli strains
| Strain/Approach | Dopamine Concentration (mg/L) | Specific Yield (mg/g biomass) | Fold Improvement |
|---|---|---|---|
| State-of-the-art (previous) | 27.0 | 5.17 | Reference |
| Knowledge-driven DBTL strain | 69.03 ± 1.2 | 34.34 ± 0.59 | 2.6× (concentration) 6.6× (specific yield) |
The implementation of the knowledge-driven DBTL approach resulted in a dopamine production strain achieving 69.03 ± 1.2 mg/L with a specific yield of 34.34 ± 0.59 mg/g biomass [26]. This represents a significant improvement over previous state-of-the-art in vivo dopamine production, with 2.6-fold higher concentration and 6.6-fold greater specific yield [26]. These dramatic improvements demonstrate the efficacy of incorporating upstream in vitro investigations to guide rational strain design before DBTL cycling.
Research Reagent Solutions
Table 2: Essential research reagents and materials for dopamine production strain development
| Reagent/Material | Function/Application | Specifications/Composition |
|---|---|---|
| HpaBC Enzyme | Converts L-tyrosine to L-DOPA | Native E. coli 4-hydroxyphenylacetate 3-monooxygenase [26] |
| Ddc Enzyme | Converts L-DOPA to dopamine | Heterologous L-DOPA decarboxylase from Pseudomonas putida [26] |
| Crude Cell Lysate System | In vitro pathway testing | E. coli lysate with metabolites, energy equivalents, and cofactors [26] |
| RBS Library Variants | Translation fine-tuning | Modified Shine-Dalgarno sequences with varying GC content [26] |
| Minimal Production Medium | Strain cultivation and evaluation | Glucose, salts, MOPS buffer, trace elements, vitamin B₆ [26] |
| Phosphate Buffer | In vitro reaction buffer | 50 mM, pH 7.0 with FeClâ‚‚ and tyrosine/DOPA substrates [26] |
The knowledge-driven DBTL framework demonstrates how upstream in vitro investigation significantly enhances the efficiency of metabolic engineering campaigns. By employing crude cell lysate systems for preliminary pathway validation and optimization, researchers can generate mechanistic insights that inform rational design decisions before resource-intensive in vivo cycling [26]. In the case study presented, this approach enabled precise RBS engineering that accounted for the impact of GC content in the Shine-Dalgarno sequence on translation efficiency, ultimately resulting in dramatically improved dopamine production strains [26].
This methodology represents an advancement in systems metabolic engineering, integrating computational design, synthetic biology tools, and automated biofoundry platforms to accelerate the development of microbial cell factories [6]. The principles established for dopamine production in E. coli are readily transferable to other valuable biochemicals, potentially transforming industrial biotechnology by providing a more efficient and mechanistic framework for optimizing complex biosynthetic pathways. Future implementations will likely incorporate more sophisticated cell-free systems and machine learning approaches to further enhance predictive design capabilities within the DBTL cycle.
The engineering of microbial cell factories for the production of fuels, chemicals, and pharmaceuticals relies on the iterative Design-Build-Test-Learn (DBTL) cycle to systematically optimize complex metabolic pathways [6]. A fundamental challenge in this process is overcoming metabolic flux imbalances that arise when pathway enzymes are expressed at non-optimal levels, leading to suboptimal product yields and the accumulation of intermediate metabolites [28]. The sequential, low-throughput testing of genetic designs represents a major bottleneck in this endeavor [29].
High-throughput toolboxes—specifically, ribosome binding site (RBS) engineering, promoter library construction, and combinatorial assembly methods—have emerged as powerful solutions to accelerate the DBTL cycle. These technologies enable the generation of vast genetic diversity and its rapid screening, facilitating the empirical discovery of optimal pathway configurations that would be difficult to predict based on first principles alone [30] [28]. By allowing researchers to perturb multiple targets in the metabolic network simultaneously and in a high-throughput manner, these toolboxes shift metabolic engineering from a sequential, rational design process to a parallel, combinatorial optimization effort [29]. This guide provides an in-depth technical examination of these core toolboxes, their integration within the DBTL framework, and the protocols that enable their application.
Toolbox 1: Promoter Engineering and Library Design
Rationale and Key Targets
Promoter libraries are essential for tuning gene expression at the transcriptional level. The need for combinatorial building and testing of promoter regions arises in various applications, including tuning gene expression, pathway optimization, designing synthetic circuits, and engineering biosensors [30]. A semi-rational approach, which targets specific functional regions of the promoter rather than randomly mutating the entire sequence, is often the most effective strategy.
Key regulatory regions for targeted mutagenesis include:
- The -35 and -10 Boxes: These regions, located approximately 35 and 10 bases upstream of the transcriptional start site, are critical for transcription initiation. Single-nucleotide mutations in these boxes can have dramatic effects on promoter strength [30].
- Operator Regions: These are palindromic or pseudo-palindromic sequences to which transcription factors bind. Randomizing a few nucleotides within operator regions can modulate the binding affinity of transcription regulators, thereby altering the dynamic response range, achieving very tight repression, or resulting in constitutive promoter activity [30].
- Ribosome Binding Sites (RBS): Although not part of the promoter itself, the RBS is frequently included in promoter library designs to enable concomitant tuning of translation initiation rates [30].
Experimental Protocol: Construction of Promoter Variant Libraries
The following protocol, adapted from high-throughput synthetic biology studies, describes the creation of promoter variant libraries using overlap extension PCR with degenerate primers [30].
Step 1: Library Design and Oligonucleotide Ordering
- Identify the target region for randomization (e.g., the -10 box, a specific operator sequence).
- Design a pair of forward and reverse primers that flank the entire promoter sequence. Within these primers, incorporate degenerate codons (e.g., NNN for complete randomization) at the targeted nucleotide positions.
- For a more focused library, use degenerate primers with reduced codon sets (e.g., NNK, where K = G or T) to lower sequence diversity while still covering all amino acids.
Step 2: Two-Round Overlap Extension PCR
- First Round (Fragment Generation): Perform separate PCRs to generate overlapping DNA fragments. Use the degenerate primers in conjunction with external forward and reverse primers to amplify the promoter regions, creating fragments with overlapping ends that contain the randomized sequence.
- Second Round (Fragment Assembly): Combine the overlapping PCR products from the first round in a subsequent PCR reaction without primers. This allows the fragments to anneal and extend via their overlapping regions. After a few cycles, add the external primers to amplify the full-length, assembled promoter variants.
Step 3: Library Cloning and Verification
- Clone the resulting PCR library into a plasmid vector upstream of a reporter gene (e.g., a fluorescent protein) using standard molecular biology techniques.
- Transform the library into the desired microbial host. The resulting library diversity can range from 10^4 to 10^7 variants [30].
- Verify the library's quality and diversity by sequencing a representative number of clones (e.g., 50-100) from the pool.
Table 1: Promoter Library Design Strategies
| Target Region | Biological Function | Mutagenesis Strategy | Expected Outcome |
|---|---|---|---|
| -35 / -10 Boxes | Transcription initiation | Complete (NNN) or partial saturation mutagenesis | Altered promoter strength (stronger/weaker constitutive activity) |
| Operator Sequence | Transcription factor binding | Saturation mutagenesis of specific DNA-binding nucleotides | Modulated induction dynamics and ligand sensitivity |
| Ribosome Binding Site | Translation initiation | Degenerate Shine-Dalgarno sequence (e.g., NNNNNNNN) | Fine-tuning of protein translation levels |
Toolbox 2: RBS Engineering for Translational Control
Principles and Computational Design
RBS engineering is a powerful method for fine-tuning the translation initiation rate (TIR) of a mRNA, thereby controlling the expression level of a specific protein without altering the promoter or the coding sequence itself [28]. A key advantage is the ability to independently adjust the expression levels of individual proteins within an operon, making it particularly valuable for balancing multi-gene pathways [28].
The challenge of naive RBS library design is combinatorial explosion. For instance, a fully degenerate 8-base Shine-Dalgarno sequence (N8) for a three-gene pathway generates 2.8 × 10^14 possible combinations, a number far beyond experimental screening capabilities [28]. Furthermore, such libraries are highly skewed, with the vast majority of sequences conferring very low TIRs.
To overcome this, algorithms like RedLibs (Reduced Libraries) have been developed [28]. RedLibs uses input from RBS prediction software (e.g., the RBS Calculator) to design a single, partially degenerate RBS sequence that yields a "smart" sub-library. This sub-library has two key features:
- It uniformly samples the entire accessible TIR space in a linear manner, ensuring coverage of low, medium, and high expression levels.
- It is condensed to a user-specified size (e.g., 4, 12, or 24 variants), making it experimentally tractable for screening.
Experimental Protocol: Implementation of a Reduced RBS Library
Step 1: In Silico Library Design with RedLibs
- Generate a gene-specific input dataset by running the RBS Calculator on a fully degenerate SD sequence for your gene of interest. This produces a list of all possible RBS sequences and their predicted TIRs.
- Input this dataset into the RedLibs algorithm along with the desired final library size.
- RedLibs performs an exhaustive search, comparing the TIR distributions of all possible partially degenerate sequences to a uniform target distribution. It returns a ranked list of optimal degenerate sequences that best match the target.
Step 2: Library Construction and Screening
- Synthesize the top degenerate RBS sequence identified by RedLibs.
- Integrate this sequence into your genetic construct via one-pot cloning, such as Golden Gate assembly or Gibson assembly, upstream of the target gene.
- Screen the resulting library. Due to its rational reduction and uniform coverage, the library will have a high density of functional clones, maximizing the chance of finding the "metabolic sweet spot" with a limited number of screened variants [28].
Step 3: Advanced Workflow: Multiplex Base Editing
- As an alternative to cloning, RBS libraries can be generated directly on the genome using CRISPR-based base editors. The bsBETTER system, for example, enables template-free, multiplexed RBS editing across many genomic loci [31].
- By co-expressing a base editor and sgRNAs targeting the RBSs of multiple genes, one can generate thousands of combinatorial variants in situ. This system has been used to edit 12 lycopene biosynthetic genes simultaneously, achieving up to 255 of 256 theoretical RBS combinations per gene and a 6.2-fold increase in lycopene production [31].
Toolbox 3: Combinatorial Pathway Assembly
The Need for Combinatorial Optimization
Even with well-characterized genetic parts, the behavior of a recombinant pathway is often unpredictable due to complex interactions and emergent regulatory mechanisms in the new host context [28]. Consequently, finding the optimal balance of absolute and relative enzyme levels requires exploring a vast multi-dimensional expression space. Combinatorial assembly methods are designed to efficiently navigate this space by generating large numbers of pathway variants in a single experiment.
Experimental Protocol: The COMPASS Method
The COMbinatorial Pathway ASSembly (COMPASS) protocol is a rapid cloning method for the balanced expression of multiple genes in a biochemical pathway [32]. It generates thousands of individual DNA constructs in a modular, parallel, and high-throughput manner.
Step 1: Modular Part Preparation
- Clone all pathway genes and regulatory elements (e.g., promoters of varying strengths, RBSs) as standardized, modular parts in compatible vectors.
Step 2: Combinatorial Assembly via Homologous Recombination
- Perform a one-pot assembly reaction where the modular parts are mixed and assembled in vivo (in yeast) or in vitro using a recombination-based cloning system.
- COMPASS employs a positive selection scheme to identify correctly assembled pathway variants, significantly reducing the background of false-positive clones.
Step 3: Multilocus Genomic Integration
- Integrate the assembled pathway variants into pre-defined loci of the host genome. COMPASS is equipped with a CRISPR/Cas9 system that enables one-step, multilocus integration of the assembled genes, ensuring stable maintenance and expression [32].
Experimental Protocol: CRISPR-AID for Combinatorial Metabolism Rewiring
For combinatorial regulation without the need for extensive DNA assembly, the CRISPR-AID system provides a powerful alternative [29]. This orthogonal tri-functional CRISPR system combines transcriptional activation (CRISPRa), transcriptional interference (CRISPRi), and gene deletion (CRISPRd) in a single plasmid.
Step 1: System Design and Construction
- Select three orthogonal CRISPR proteins (e.g., dSpCas9 for activation, dSt1Cas9 for interference, and LbCpf1 for deletion).
- Fuse the nuclease-deficient Cas proteins to appropriate effector domains (e.g., VPR for activation, MXII for repression).
- Clone expression cassettes for these proteins and their respective guide RNAs (gRNAs) targeting your pathway genes of interest into a single plasmid.
Step 2: Library Generation and Screening
- Transform the CRISPR-AID plasmid library into your production host. The simultaneous expression of multiple gRNAs will generate a population of cells with diverse genetic perturbations.
- Screen or select for clones with improved product yield. In a case study, this approach achieved a 3-fold increase in β-carotene production in S. cerevisiae in a single step [29].
Integration within the DBTL Cycle and Workflow Visualization
The true power of these toolboxes is realized when they are seamlessly integrated into the DBTL cycle. The trend is shifting towards a data-driven LDBT cycle, where Machine Learning (ML) and prior knowledge guide the initial design, accelerating the entire process [5].
- Learn → Design: Pre-trained protein language models (e.g., ESM, ProGen) and biophysical models can be used for zero-shot design of optimized sequences, or algorithms like RedLibs can design optimally diverse and compact DNA libraries for experimental testing [28] [5].
- Build: High-throughput DNA synthesis and assembly services, automated biofoundries, and cell-free expression systems accelerate the construction of genetic variants. Cell-free systems are especially powerful for rapid, high-yield protein synthesis without cloning [5].
- Test: Fluorescence-activated cell sorting (FACS) enables the ultra-high-throughput screening of cellular libraries based on fluorescent reporters [30]. For cell-free expressed libraries, droplet microfluidics can screen hundreds of thousands of reactions [5].
- Learn: Data from the Test phase are used to train ML models, which in turn generate improved designs for the next DBTL cycle, creating a virtuous cycle of optimization [5].
The following diagram illustrates the integrated high-throughput workflow for combinatorial pathway optimization, from library design to hit isolation.
Essential Research Reagent Solutions
The successful implementation of these toolboxes relies on a suite of essential reagents and platforms. The table below details key solutions for constructing and screening combinatorial libraries.
Table 2: Key Research Reagent Solutions for High-Throughput Toolboxes
| Reagent / Platform | Supplier / Source | Function in Workflow |
|---|---|---|
| Degenerate Oligonucleotides | Commercial DNA Synthesis Companies | Introduces controlled diversity at specific nucleotide positions during PCR for library generation [30]. |
| Overlap Extension PCR Reagents | Standard Molecular Biology Suppliers | Enables two-step assembly of gene or promoter variant libraries from smaller fragments [30]. |
| Cell-Free Protein Expression System | Purified Components or Crude Lysates | Provides a rapid, high-throughput platform for protein synthesis and testing without live cells, enabling megascale data generation [5]. |
| Fluorescence-Activated Cell Sorter (FACS) | Flow Cytometry Core Facilities | Enables ultra-high-throughput screening and isolation of rare library variants based on fluorescent reporters [30]. |
| CRISPR Protein Orthologs (SpCas9, SaCas9, LbCpf1) | Academic Cloning Resources / Addgene | Forms the core of orthogonal multi-functional systems for simultaneous activation, interference, and deletion (CRISPR-AID) [29]. |
| RBS Calculator & RedLibs Algorithm | Publicly Available Software | Computational tools for predicting translation initiation rates and designing optimally reduced RBS libraries [28]. |
RBS engineering, promoter libraries, and combinatorial assembly are no longer niche techniques but are central to modern, high-throughput metabolic engineering. By enabling the systematic and parallel exploration of a vast genetic design space, these toolboxes directly address the core challenge of the DBTL cycle: the efficient optimization of complex biological systems. The integration of these experimental toolboxes with computational design and machine learning is paving the way for a new paradigm of biological engineering, moving from iterative trial-and-error towards more predictive and rational design.
Integrating Cell-Free Systems for Rapid Building and Testing
The Design-Build-Test-Learn (DBTL) cycle has long been the cornerstone of metabolic engineering and synthetic biology, providing a systematic framework for engineering biological systems. Traditionally, this iterative process begins with Design, followed by physical implementation (Build), experimental validation (Test), and data analysis to inform the next cycle (Learn). However, recent technological advancements are fundamentally reshaping this paradigm. The integration of machine learning (ML) and cell-free systems is accelerating DBTL cycles, enabling a proposed reordering to LDBT (Learn-Design-Build-Test) where learning precedes design [5] [33].
This whitepaper provides an in-depth technical guide on integrating cell-free protein synthesis (CFPS) platforms into metabolic engineering research. CFPS systems leverage the protein biosynthesis machinery from crude cell lysates or purified components to activate in vitro transcription and translation [5]. By decoupling complex biological functions from the constraints of living cells, these systems offer unprecedented control and speed for the Build and Test phases. When combined with machine learning-driven design, they facilitate a powerful, closed-loop engineering pipeline capable of dramatically reducing development timelines from months to days [34] [33].
The LDBT Framework: A Learn-First Approach
Core Principles and Workflow
The LDBT framework initiates with a data-driven learning phase, where machine learning models pre-trained on vast biological datasets generate predictive hypotheses. This is followed by computational Design, rapid Build using cell-free systems, and high-throughput Testing whose outcomes further enrich the learning repository [5] [33]. This reordering leverages the power of zero-shot predictions—where models make accurate functional forecasts without additional training—allowing researchers to start from a more informed design space and potentially achieve functional systems in a single cycle [5].
Key computational tools enabling this shift include:
- Sequence-based models like ESM [35] and ProGen [36] trained on evolutionary relationships in protein sequences.
- Structure-based tools like ProteinMPNN [37] and MutCompute [38] for designing sequences that fold into desired structures or optimizing residues based on local environment.
- Hybrid approaches combining evolutionary information with biophysical principles for enhanced predictive power [5] [35].
The diagram below illustrates the core workflow and logical relationships of the integrated LDBT framework with cell-free systems.
Quantitative Performance of Integrated LDBT Systems
The synergy between machine learning and cell-free testing delivers measurable improvements in optimization efficiency. The following table summarizes key performance metrics from recent implementations.
Table 1: Quantitative Performance of Integrated LDBT Systems
| Application | Experimental Rounds | Performance Improvement | Key Enabling Technologies | Reference |
|---|---|---|---|---|
| Colicin M & E1 Production | 4 DBTL cycles | 2- to 9-fold yield increase | Active Learning with Cluster Margin sampling; Automated ChatGPT-4 generated code | [34] |
| PET Hydrolase Engineering | Not specified | Increased stability and activity vs. wild-type | MutCompute structure-based deep neural network | [5] |
| TEV Protease Design | Not specified | ~10x increase in design success rates | ProteinMPNN + AlphaFold/RoseTTAFold structure assessment | [5] |
| Antimicrobial Peptide Screening | Computational survey of >500,000 variants | 6 promising AMP designs validated | Deep-learning sequence generation + cell-free expression | [5] |
| 3-HB Biosynthesis | Not specified | >20-fold improvement in yield | iPROBE (neural network-guided pathway optimization) | [5] |
Technical Implementation: Cell-Free System Methodologies
Core Reagent Solutions for CFPS
Cell-free systems utilize carefully formulated reagent mixtures to support transcription and translation in vitro. The table below details essential components and their functions.
Table 2: Key Research Reagent Solutions for Cell-Free Protein Synthesis
| Reagent Category | Specific Components | Function in CFPS | Technical Notes |
|---|---|---|---|
| Energy Source | Phosphoenolpyruvate (PEP), Creatine Phosphate, Glucose | Drives ATP regeneration for transcription/translation | Recent formulations include ribose and starch as accessory substrates [39] |
| Cell Extract | E. coli lysate, HeLa lysate, PURExpress | Provides transcriptional/translational machinery & core metabolism | Extract origin determines PTM capability; eukaryotic extracts enable native glycosylation [34] [39] |
| Amino Acid Mixture | 20 standard amino acids | Building blocks for protein synthesis | Enables incorporation of non-canonical amino acids for expanded functionality [5] |
| Nucleotide Mix | ATP, GTP, CTP, UTP | Substrates for RNA polymerase during transcription | Maintains proper NTP ratios to prevent transcriptional stalling |
| Cofactors | Mg²âº, Kâº, cyclic AMP | Essential enzyme cofactors for energy metabolism & polymerases | Concentration optimization critical; affects folding and activity [34] |
| DNA Template | PCR product or plasmid | Genetic blueprint for target protein | No cloning required; direct use of synthesized DNA significantly accelerates Build phase [5] |
Experimental Protocol: Automated DBTL for CFPS Optimization
The following detailed protocol is adapted from a fully automated DBTL pipeline for optimizing colicin production in CFPS systems [34].
Phase 1: Design (Automated Computational Design)
- Objective Definition: Specify target protein and optimization goals (e.g., yield, solubility, activity).
- Template Preparation: Generate DNA template sequences via PCR or synthetic gene fragments.
- Experimental Design Generation: Use automated scripts (Python) to create a Design of Experiments (DoE) matrix. This defines the combinations of CFPS components to test.
- Technical Insight: This phase was implemented using ChatGPT-4 generated code without manual revision, demonstrating the accessibility of automated design for non-programmers [34].
Phase 2: Build (Automated Reaction Assembly)
- Master Mix Preparation: Combine in a 1.5 mL microtube:
- 70% (vol/vol) cell extract (E. coli or HeLa, clarified by centrifugation at 12,000 × g for 10 min)
- Energy Solution: 2 mM ATP, 2 mM GTP, 1 mM CTP, 1 mM UTP
- Amino Acid Mix: 1 mM of each standard amino acid
- Energy Regeneration: 20 mM phosphoenolpyruvate
- Cofactors: 1.5 mM Mg²âº, 80 mM Kâº, 50 mM HEPES buffer (pH 7.6)
- Aliquoting: Using a liquid handling robot, dispense 10 µL of master mix into each well of a 96-well microplate.
- Template Addition: Add 1 µL of DNA template (50-100 ng/µL) to respective wells as per the DoE layout.
- Incubation: Seal plate and incubate at 30-37°C for 4-6 hours with orbital shaking (300 rpm) for aeration.
Phase 3: Test (High-Throughput Analysis)
- Yield Quantification:
- Measure absorbance at 600 nm (turbidity) or use fluorescence-based assays (e.g., GFP-fusion).
- Apply colorimetric assays (Bradford/Lowry) against a BSA standard curve.
- Functional Assessment:
- For colicins: Perform antimicrobial activity assays against sensitive E. coli strains via zone-of-inhibition [34].
- For enzymes: Implement coupled enzyme assays or HPLC for metabolite detection.
Phase 4: Learn (Data Analysis & Active Learning)
- Data Processing: Normalize and preprocess test data (e.g., expression yields, activity metrics).
- Model Training: Employ machine learning (e.g., random forest, neural networks) to map CFPS composition to target outcomes.
- Candidate Selection: Apply Active Learning with Cluster Margin sampling to identify the most informative subsequent experiments balancing uncertainty and diversity [34].
- Cycle Iteration: Feed selected candidates into the next automated Design phase.
The automated workflow integrating these phases is depicted below.
Advanced Applications in Metabolic Engineering
Pathway Prototyping and Optimization
Cell-free systems excel at rapid pathway prototyping by combining multiple enzymes in controlled ratios. The iPROBE (in vitro prototyping and rapid optimization of biosynthetic enzymes) platform uses neural networks to predict optimal pathway sets and enzyme expression levels from training data of pathway combinations [5]. This approach achieved over 20-fold improvement in 3-HB production in Clostridium [5]. A key advantage is the ability to test non-native pathways in a decoupled environment, bypassing host-specific issues like toxicity, regulatory networks, and resource competition [39].
Expanding Metabolic Capabilities
CFPS platforms are expanding beyond traditional pathways to incorporate:
- C1 Substrate Conversions: Utilizing cell extracts from engineered strains to activate pathways for formate, methanol, and COâ‚‚ valorization [39].
- Non-Model Organism Metabolism: Leveraging extracts from diverse organisms (including extremophiles) to access unique biochemical capabilities [39].
- Hybrid Systems: Mixing extracts from multiple organisms (e.g., cyanobacteria and E. coli) to combine metabolic functions not found in a single species [39].
Implementation Considerations
Integration with Biofoundries
Biofoundries provide the ideal infrastructure for implementing integrated LDBT cycles with cell-free systems. These facilities combine automation, robotic liquid handling, and bioinformatics to streamline synthetic biology workflows [40]. The Global Biofoundry Alliance now includes over 30 members worldwide, standardizing development efforts through initiatives like SynBiopython [40].
Technical Challenges and Solutions
- Data Quality and Quantity: ML models require large, high-quality datasets. Solution: Leverage ultra-high-throughput screening like droplet microfluidics (e.g., DropAI screening 100,000+ reactions) [5].
- System Robustness: Cell-free reactions can be sensitive to component variations. Solution: Implement rigorous quality control for cell extract preparation and reagent formulation.
- Cost Management: Scale-dependent costs can be prohibitive. Solution: Optimize miniaturization and leverage open-source automation platforms like AssemblyTron [40].
- Predictive Model Accuracy: Biological complexity challenges prediction. Solution: Combine multiple modeling approaches (sequence, structure, biophysics) and continuously refine with experimental data [5].
The integration of cell-free systems for rapid building and testing within the DBTL framework represents a transformative advancement for metabolic engineering research. By combining the predictive power of machine learning with the experimental agility of cell-free platforms, researchers can dramatically accelerate the development of novel biosynthetic pathways and engineered proteins. The emerging LDBT paradigm—where learning precedes design—promises to reshape synthetic biology, moving the field closer to a predictive engineering discipline capable of solving complex challenges in biomanufacturing, therapeutic development, and sustainable bio-production.
Overcoming Bottlenecks: Strategies for Efficient and Robust DBTL Cycling
In the structured framework of the Design-Build-Test-Learn (DBTL) cycle for metabolic engineering, the Build phase is a critical translational step. It is where designed genetic constructs are physically synthesized and assembled into host organisms [41]. This phase has traditionally been a major bottleneck, often consuming significant time and resources, thereby slowing the iterative pace essential for rapid strain development [42] [3]. The efficiency of the entire DBTL cycle is heavily dependent on the speed, accuracy, and cost-effectiveness of building genetic variants.
This whitepaper addresses two pivotal hurdles within the Build phase: DNA synthesis and automated clone selection. We provide an in-depth technical analysis of current technologies, detailed protocols, and emerging trends aimed at empowering researchers and drug development professionals to accelerate their metabolic engineering workflows.
DNA Synthesis: From Oligos to Genes
DNA synthesis is the foundational process of artificially creating DNA sequences de novo, providing the raw genetic material for pathway engineering [43]. The field has evolved significantly from manual, low-throughput methods to highly automated and scalable services.
DNA Synthesis Technologies and Market Landscape
The global DNA synthesis market, valued at USD 4.56 billion in 2024, is projected to grow to USD 16.08 billion by 2032, exhibiting a compound annual growth rate (CAGR) of 17.5% [43]. This growth is propelled by rising demand from the pharmaceutical and biotechnology sectors. The market is segmented by type into oligonucleotide synthesis and gene synthesis, with the latter expected to retain the largest market share due to its customizable applications for research and therapeutics [43].
Table 1: Key DNA Synthesis Technologies and Commercial Providers
| Technology | Key Principle | Advantages | Limitations/Challenges | Representative Companies |
|---|---|---|---|---|
| Traditional Solid-Phase Synthesis [44] | Step-by-step chemical synthesis (phosphoramidite method) on a solid support. | Mature, reliable process; high precision for target sequences. | Low synthesis efficiency; complex operation; high cost for large-scale use. | Gene Art (Thermo Fisher Scientific) |
| Chip-Based High-Throughput Synthesis [44] | Uses silicon chips to synthesize thousands of oligonucleotides in parallel. | Dramatically increased throughput; lower cost per sequence; high flexibility. | Lower accuracy for complex sequences (e.g., high GC, repeats). | Twist Bioscience |
| AI-Powered Gene Synthesis [44] | AI algorithms analyze and optimize gene sequences prior to synthesis. | Improves synthesis success for complex sequences; enables codon optimization for expression. | AI algorithms are still under development and require continuous optimization. | Synbio Technologies |
| Enzymatic DNA Synthesis [45] | Uses terminal deoxynucleotidyl transferase (TdT) enzymes to assemble DNA. | Potentially greener (less waste); can generate longer and more accurate sequences. | Emerging technology; requires refinement for widespread commercial use. | Ansa Biotechnologies |
A key trend is the industry's focus on developing faster, cheaper, and more accurate synthesis methods [43]. For instance, microfluidics drastically reduces turnaround time via an automated system that enhances the parallelization of reactions [43]. Furthermore, enzyme-based DNA synthesis is emerging as a sustainable alternative to traditional chemical methods, showing promise for generating extended, high-fidelity DNA sequences [45].
Experimental Workflow for High-Throughput Gene Synthesis
The following diagram illustrates a generalized workflow for constructing a gene library, integrating both high-throughput synthesis and subsequent clone selection.
Protocol: Chip-Based Gene Library Synthesis and Assembly [44] [46]
- Design: The target gene sequence is designed in silico. For metabolic pathway optimization, this often involves creating a library of variants with different promoters, ribosome binding sites (RBS), or codon-optimized coding sequences. AI tools can be employed here to predict and optimize sequences for high expression and stability [44].
- Oligonucleotide Synthesis: Thousands of unique oligonucleotides (typically 60-200 nt) are synthesized in parallel on a silicon chip using proprietary high-throughput phosphoramidite chemistry [44].
- Oligo Pool Recovery: The synthesized oligos are cleaved from the chip surface and pooled.
- Gene Assembly: The oligonucleotide pools are assembled into full-length genes. Common methods include:
- PCR-Based Assembly: Overlapping oligos are used as templates in a polymerase chain reaction to assemble the full gene.
- Enzymatic Assembly: Using methods like Golden Gate assembly, which leverages Type IIS restriction enzymes for seamless, scarless assembly of multiple DNA fragments [42].
- Cloning: The assembled genes are cloned into a plasmid vector via automated liquid handling workstations, which perform restriction digestion, ligation, and other enzymatic reactions [46].
- Transformation: The constructed plasmids are introduced into a microbial host (e.g., E. coli) for propagation.
Automated Clone Selection: Isolating the Right Construct
Following DNA assembly and transformation, the next critical hurdle is efficiently isolating correctly constructed clones from a background of empty vectors or incorrectly assembled products. Traditional manual colony picking is time-consuming and a significant bottleneck in high-throughput workflows [42].
Automated Clone Selection Methods
Table 2: Comparison of Clone Selection Methods
| Method | Principle | Throughput | Selectivity | Cost & Complexity |
|---|---|---|---|---|
| Manual Colony Picking [42] | Visual identification and manual transfer of individual colonies from agar plates. | Low | High (if performed carefully) | Low equipment cost, but high labor time. |
| Automated Colony Picking Robots [42] | Robotic arms with vision systems to identify and pick colonies from agar plates. | High | Can be hampered by overlapping colonies or uneven agar [42] | High initial investment; complex setup. |
| Flow Cytometry with Biosensors [42] | Cell sorting based on fluorescence activated by biosensors reporting on specific pathways. | High | High (if a specific biosensor is available) | Moderate to high; requires biosensor engineering and FACS equipment. |
| Automated Liquid Clone Selection (ALCS) [42] | Exploits uniform growth behavior of correctly transformed cells in liquid culture under selective pressure. | High | Reported 98 ± 0.2% for correctly transformed E. coli [42] | Low; requires only standard liquid handling robotics. |
For academic or semi-automated biofoundries, the Automated Liquid Clone Selection (ALCS) method presents a robust and cost-effective alternative to sophisticated colony-picking robotics [42]. This "low-tech" method achieves high selectivity by leveraging the uniform growth kinetics of successfully transformed cells in liquid media, without requiring additional capital investment.
Experimental Protocol: Automated Liquid Clone Selection (ALCS)
The ALCS method is particularly well-suited for organisms like E. coli, Pseudomonas putida, and Corynebacterium glutamicum [42]. The workflow is visualized below.
Detailed ALCS Protocol for E. coli [42]:
- Transformation: Perform a standard transformation of the assembled plasmid DNA into a competent host strain (e.g., E. coli BL21(DE3)).
- Outgrowth: Add transformation mixture to SOC medium and incubate with shaking for 1 hour at 37°C.
- Dilution and Dispensing: Dilute the outgrowth culture in a selective liquid medium (e.g., 2xTY with appropriate antibiotic) to a target low cell density. Using an automated liquid handler, dispense the diluted culture into a 96-well or 384-well microtiter plate.
- Cultivation and Monitoring: Incubate the plate with shaking at 37°C while monitoring optical density (OD600). The incubation should cover approximately five generations to allow for clear growth differentiation.
- Model-Based Selection: Correctly transformed cells will exhibit uniform and robust growth. A model-based setup analyzes growth curves and selects only those cultures that exceed a predefined growth threshold, indicating the presence of the antibiotic resistance gene and successful transformation. This step achieves the reported high selectivity of 98% [42].
- Downstream Processing: The selected cultures can be used directly for subsequent steps in the DBTL cycle, such as inoculating test-phase cultures for metabolite screening.
The Scientist's Toolkit: Essential Research Reagents and Materials
Table 3: Key Reagent Solutions for DNA Synthesis and Clone Selection
| Item | Function/Application | Example Use Case |
|---|---|---|
| Silicon DNA Synthesis Chip [44] | Platform for parallel synthesis of thousands of oligonucleotides. | High-throughput gene library synthesis by Twist Bioscience. |
| Golden Gate Assembly Mix [42] | Enzyme mix (Type IIS restriction enzyme + ligase) for seamless, one-pot DNA assembly. | Robust and efficient assembly of multiple DNA fragments with high cloning efficiency. |
| Chemically Competent Cells [42] | Bacterial cells treated for efficient plasmid uptake via heat shock. | Routine cloning in E. coli DH5α for plasmid propagation. |
| Electrocompetent Cells [42] | Bacterial cells prepared for efficient plasmid uptake via electroporation. | Transformation of large plasmids or less efficient strains like C. glutamicum. |
| Selective Liquid Media [42] [3] | Growth media containing antibiotics to select for successfully transformed clones. | Essential for ALCS protocol (e.g., 2xTY + ampicillin). |
| SOC Outgrowth Medium [42] [3] | Nutrient-rich recovery medium for cells after transformation. | Allows for expression of antibiotic resistance genes before plating or liquid selection. |
| Ammonium sulfide ((NH4)2(S5)) | Ammonium Sulfide ((NH4)2(S5))|178.4 g/mol|RUO | |
| 4,8,12-Trioxatridecan-1-ol | 4,8,12-Trioxatridecan-1-ol | CAS 13133-29-4 | 4,8,12-Trioxatridecan-1-ol (C10H22O4) is a polyether alcohol for use as a hydrophilic linker in drug discovery and materials science. For Research Use Only. Not for human or veterinary use. |
The Build phase is undergoing a profound transformation driven by automation, data science, and novel biochemistry. Key future trends include:
- The Rise of AI and "LDBT" Cycles: Machine learning is shifting the paradigm from Design-Build-Test-Learn (DBTL) to Learn-Design-Build-Test (LDBT). In this model, AI models pre-trained on vast biological datasets are used for zero-shot design, potentially generating functional genetic constructs from the outset, which are then rapidly built and tested [5].
- Integration of Cell-Free Systems: Cell-free gene expression platforms are accelerating the Build and Test phases by enabling rapid, high-throughput protein and pathway prototyping without the constraints of living cells, seamlessly integrating with AI-driven design [5].
- In Vivo Synthesis: A longer-term vision involves using engineered cells as "factories" to synthesize target DNA sequences directly in vivo, leveraging the cell's own replication and repair machinery for high-fidelity, scalable production [44].
In conclusion, overcoming the critical Build-phase hurdles of DNA synthesis and clone selection is paramount for accelerating metabolic engineering. By leveraging high-throughput, automated DNA synthesis technologies and implementing cost-effective, robust methods like Automated Liquid Clone Selection, researchers can significantly streamline their DBTL cycles. This enhanced capability enables more rapid iteration and optimization, ultimately speeding up the development of novel microbial cell factories for therapeutic, chemical, and biofuel production.
Leveraging Machine Learning for Predictive Modeling in Low-Data Regimes
The Design-Build-Test-Learn (DBTL) cycle is a foundational framework in metabolic engineering for the systematic development of microbial cell factories [47]. However, its effectiveness has traditionally been hampered in the Learn phase, where limited experimental data often restricts the development of predictive models [2] [48]. The challenge of operating in low-data regimes is a significant bottleneck, making the building of reliable models difficult and often forcing researchers to rely on intuition. This technical guide explores how machine learning (ML) can be integrated with mechanistic models and strategic experimental design to create powerful predictive capabilities even when data is scarce, thereby accelerating the optimization of metabolic pathways for the production of valuable compounds.
The Data Challenge in Metabolic Engineering DBTL Cycles
In a typical DBTL cycle, the Build and Test phases generate the data that feeds the Learn phase, where computational models are developed to inform the next Design phase [10]. The core challenge in the Learn phase is the limited quantity of high-quality experimental data that can be feasibly generated. This creates a significant capability gap, where the capacity to design and build genetic constructs far outpaces the ability to test and learn from them [2].
High-throughput analytics can generate large datasets, but they are often constrained by cost, time, and technical limitations. Consequently, learning is frequently the most weakly supported step in the cycle [2]. Machine learning models, which are inherently data-hungry, face particular difficulties in this environment. Without ample training data, their predictions can be unreliable, a problem often termed "garbage in, garbage out" [49]. Overcoming this requires innovative strategies that make the most of every data point.
Integrated Frameworks for Low-Data Regimes
Combining Mechanistic and Machine Learning Models
A powerful strategy to overcome data scarcity is the integration of mechanistic models with data-driven ML approaches. Mechanistic models, such as Genome-Scale Metabolic Models (GSMMs), are built on prior knowledge of the stoichiometry and structure of metabolic networks [50] [51]. While they may lack the predictive power for complex phenotypes on their own, they can be used to generate informative in-silico data or to identify the most promising regions of the vast genetic design space for experimental testing [50] [51].
This hybrid approach was successfully demonstrated for engineering aromatic amino acid metabolism in yeast. A genome-scale model first pinpointed potential gene targets. A subset of these targets was then used to construct a combinatorial library. The experimental data from testing this library was used to train various ML algorithms, which subsequently recommended designs that improved tryptophan titer by up to 74% compared to the best designs used in the training set [50]. This exemplifies how a mechanistic model can guide efficient library construction, ensuring that the limited experimental data collected is of high value for training the ML model.
The Automated Recommendation Tool (ART)
The Automated Recommendation Tool (ART) is a machine learning solution specifically designed for the constraints of synthetic biology, including low-data regimes [48]. ART leverages a Bayesian ensemble approach to provide probabilistic predictions. Instead of giving a single, potentially overfitted output, it provides a distribution of possible outcomes, quantifying the uncertainty in its predictions [48]. This is crucial for guiding experimental design, as it allows researchers to balance the exploration of uncertain regions of the design space with the exploitation of known high-performing areas.
ART is designed to function effectively with the small datasets (often <100 instances) typical of metabolic engineering projects. It has been validated in several real-world applications, including guiding the improvement of tryptophan productivity in yeast by 106% from a base strain [48].
Knowledge-Driven DBTL and In Vitro Prototyping
Another strategy to maximize learning from limited data is the knowledge-driven DBTL cycle. This approach uses upstream in vitro investigations to gain mechanistic insights and pre-validate designs before committing to more resource-intensive in vivo strain construction [3].
For instance, in developing an E. coli strain for dopamine production, researchers first used cell-free protein synthesis (CFPS) systems to test different relative enzyme expression levels in a bi-cistronic pathway [3]. This in vitro prototyping provided high-quality data on pathway bottlenecks and optimal expression ratios without the complexities of the living cell. This knowledge was then used to design a focused in vivo library for fine-tuning via RBS engineering, resulting in a 2.6 to 6.6-fold improvement over state-of-the-art production levels [3]. This method ensures that every in vivo experiment is highly informed, dramatically increasing the learning efficiency per cycle.
Key Machine Learning Techniques and Data Integration Methods
Machine Learning Algorithms for Small Datasets
In low-data regimes, the choice of ML algorithm is critical. Complex, high-capacity models like deep learning are prone to overfitting and are generally not suitable. Instead, the following are often more effective [48] [51]:
- Bayesian Models: Provide uncertainty estimates and are robust with limited data.
- Support Vector Machines (SVMs): Effective for classification and regression in high-dimensional spaces.
- Tree-Based Methods (e.g., Random Forests): Offer good interpretability and performance.
These models are typically used within a supervised learning framework to solve regression (e.g., predicting titer) or classification (e.g., identifying high-producing strains) problems [51].
Multi-Omic Data Integration
To enrich limited datasets, ML can be applied to integrated multi-omic data (genomics, transcriptomics, proteomics, metabolomics). Integration methods include [51]:
- Concatenation-based (Early Integration): Fusing multiple data matrices into a single comprehensive matrix after normalization.
- Transformation-based (Intermediate Integration): Converting datasets into an intermediate form like a kernel matrix before integration.
- Model-based (Late Integration): Combining predictions from models trained on individual data types.
This multiview approach merges experimental data with knowledge-driven data (e.g., fluxomic data generated from GSMMs), incorporating key mechanistic information into an otherwise biology-agnostic learning process [51].
Experimental Protocols for Data Generation in Low-Data Regimes
Protocol: High-Throughput Biosensor-Based Screening for ML Training
This protocol was used to generate data for training ML models in optimizing tryptophan production in yeast [50].
- Strain Design: Construct a combinatorial library of strain variants. For example, a library of 7776 designs was created by combining five genes (e.g., CDC19, TKL1, TAL1, PCK1, PFK1) with six different promoters each [50].
- Biosensor Integration: Engineer a tryptophan biosensor into the host platform strain. This biosensor produces a fluorescent output correlated with intracellular tryptophan concentration [50] [2].
- Cultivation and Measurement: Grow library variants in 96-deepwell plates. Monitor growth and biosensor fluorescence over time using a plate reader [50].
- Data Extraction: From the time-series data, calculate the fluorescence synthesis rate (a proxy for tryptophan production rate) for each strain variant. This high-throughput method allowed the collection of >124,000 data points from ~250 strains, providing a rich dataset for ML training without requiring chromatographic analysis of every sample [50].
Protocol: In Vitro Pathway Prototyping with Cell-Free Systems
This protocol is used for knowledge-driven DBTL to gain preliminary data with minimal resource investment [3].
- Cell-Free Lysate Preparation: Cultivate the production host (e.g., E. coli), harvest cells during mid-log phase, and prepare a crude cell lysate to supply metabolites, energy equivalents, and the transcription/translation machinery [3].
- In Vitro Transcription/Translation: For each pathway variant (e.g., with different RBS sequences), set up a reaction mixture containing the cell lysate, plasmid DNA encoding the pathway genes, and necessary substrates (e.g., L-tyrosine for dopamine production) [3].
- Product Quantification: After incubation, quantify the target product and key intermediates using fast ultra-performance liquid chromatography coupled to tandem mass spectrometry (UPLC-MS/MS) [3].
- Data Analysis: Identify the relative enzyme expression levels that minimize intermediate accumulation and maximize product yield. This data directly informs the design of the subsequent in vivo library.
Visualization of Workflows
Hybrid Modeling DBTL Workflow
The following diagram illustrates the integrated DBTL cycle where mechanistic models enhance the learning phase in a low-data regime.
Knowledge-Driven DBTL with In Vitro Prototyping
This diagram outlines the workflow for a knowledge-driven DBTL cycle that uses cell-free systems to de-risk the initial learning phase.
Quantitative Results and Performance
Table 1: Performance of ML-Guided Metabolic Engineering in Low-Data Regimes
| Target Compound | Host Organism | Key Strategy | Dataset Size for Training | Reported Improvement | Citation |
|---|---|---|---|---|---|
| Tryptophan | S. cerevisiae (Yeast) | GSM + Combinatorial Library + ML | ~250 strains (from 7776 design space) | Up to 74% increase in titer | [50] |
| Tryptophan | S. cerevisiae (Yeast) | Automated Recommendation Tool (ART) | Not Specified | 106% improvement from base strain | [48] |
| Dopamine | E. coli | Knowledge-Driven DBTL (In Vitro Prototyping) | In vitro data informed in vivo library | 2.6 to 6.6-fold over state-of-the-art | [3] |
| (2S)-Pinocembrin | E. coli | Automated DBTL with DoE | 16 constructs (from 2592 design space) | 500-fold increase from initial design | [10] |
Table 2: Comparison of Machine Learning Techniques for Low-Data Applications
| ML Technique | Key Characteristics | Advantages for Low-Data Regimes | Example Use Case |
|---|---|---|---|
| Bayesian Ensemble Models (e.g., in ART) | Provides probabilistic predictions and uncertainty quantification. | Guides exploration; robust to overfitting; ideal for recommendation. | Recommending high-producing strain designs [48]. |
| Support Vector Machines (SVM) | Finds optimal hyperplane for classification/regression in high-dimensional space. | Effective with a clear margin of separation; memory efficient. | Classifying strain performance based on proteomic profiles [51]. |
| Tree-Based Methods (e.g., Random Forests) | Ensemble of decision trees; offers feature importance. | Good interpretability; handles mixed data types; reduces variance. | Identifying key genetic promoters influencing production [50]. |
| Multi-Omic Integration | Combines data from multiple sources (genomics, proteomics, etc.). | Enriches dataset; provides a more complete systems view. | Predicting phenotype from integrated transcriptomic and fluxomic data [51]. |
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Research Reagents and Platforms
| Reagent / Platform | Function | Role in Low-Data ML |
|---|---|---|
| Genome-Scale Metabolic Model (GSMM) | A knowledge-driven computational model of an organism's metabolism. | Provides priors and generates in-silico data to guide initial library design, making experimental data more informative [50] [51]. |
| Automated Recommendation Tool (ART) | A machine learning tool that uses Bayesian modeling to recommend strains. | Specifically designed for small datasets in synthetic biology; quantifies prediction uncertainty to guide the next DBTL cycle [48]. |
| Cell-Free Protein Synthesis (CFPS) System | A crude cell lysate that supports in vitro transcription and translation. | Enables high-throughput, low-cost prototyping of pathway variants to generate initial high-quality data for learning [3]. |
| Metabolite Biosensors | Genetic circuits that produce a measurable output (e.g., fluorescence) in response to a target metabolite. | Allows high-throughput, real-time monitoring of production in living cells, generating large-scale dynamic data for ML from a limited number of strains [50] [2]. |
| Design of Experiments (DoE) | A statistical method for planning experiments to maximize information gain. | Reduces the number of builds required to effectively explore a large combinatorial design space, creating optimal small datasets for ML [10]. |
| cis-2,5-Dimethyl-3-hexene | cis-2,5-Dimethyl-3-hexene, CAS:10557-44-5, MF:C8H16, MW:112.21 g/mol | Chemical Reagent |
| 3-Hydroxymethylaminopyrine | 3-Hydroxymethylaminopyrine, CAS:13097-17-1, MF:C13H17N3O2, MW:247.29 g/mol | Chemical Reagent |
The Design-Build-Test-Learn (DBTL) cycle is a cornerstone of modern metabolic engineering, enabling the systematic development of microbial cell factories for sustainable bioproduction [6]. This iterative process integrates computational design and experimental validation to optimize complex biological systems. However, its efficacy is often compromised by underlying challenges in three critical areas: data set biases, experimental noise, and library design. These factors can significantly impede cycle efficiency, leading to suboptimal strains and prolonged development timelines.
This technical guide provides an in-depth analysis of these core challenges, framed within the context of advancing systems metabolic engineering. We detail methodological frameworks for identifying, quantifying, and mitigating these issues, supported by structured data presentation and executable experimental protocols. The objective is to equip researchers with the strategies necessary to enhance the robustness, predictability, and throughput of their DBTL operations, thereby accelerating the development of next-generation bacterial cell factories [8].
The DBTL Cycle in Modern Metabolic Engineering
The DBTL cycle represents an iterative framework for strain engineering. In the Design phase, metabolic models and genetic blueprints are created. The Build phase involves the physical construction of genetic variants in a host organism like Corynebacterium glutamicum. The Test phase characterizes the performance of these constructs, and the Learn phase uses the resulting data to inform the next design iteration [6].
Recent advances have focused on increasing the throughput and automation of the entire cycle. The integration of synthetic biology (SynBio), automation, and artificial intelligence (AI) and machine learning (ML) is revolutionizing the process, enabling the development of sophisticated biodesign automation (BDA) platforms [8]. This evolution from traditional metabolic engineering to systems metabolic engineering leverages omics data, enzyme engineering, and evolutionary strategies within the DBTL framework to optimize metabolic pathways comprehensively [6].
The following diagram illustrates the core DBTL cycle and the key challenges at each stage that will be discussed in this guide.
DBTL Cycle with Key Challenges
Data Set Biases: Identification and Mitigation
Data set biases introduce systematic errors that can misdirect the learning phase, causing successive DBTL cycles to converge on local optima rather than globally superior solutions. These biases often originate from historical data, analytical instrument limitations, and sample processing protocols.
Quantitative Framework for Bias Assessment
Table 1: Common Data Set Biases and Their Impact on DBTL Learning
| Bias Category | Origin in DBTL Cycle | Impact on Model Training | Detection Method |
|---|---|---|---|
| Selection Bias | Test-phase analysis limited to high-producing clones, ignoring low-performers and failures. | Trained models lack information on non-productive genetic regions, reducing predictive range. | Analyze clone selection criteria; compare distribution of selected vs. constructed variants. |
| Measurement Bias | Analytical instruments (e.g., HPLC, MS) with non-linear response outside calibration range. | Quantitative relationships between pathway flux and product titer are distorted. | Standard reference materials and spike-in controls; assess instrument calibration curves. |
| Confounding Bias | Batch effects in Test phase from culture condition variations (pH, DO, nutrient depletion). | Model incorrectly attributes performance variation to genetic design instead of experimental artifact. | Randomized block experiments; mixed-effects statistical models to account for batch effects. |
Experimental Protocol for Bias Mitigation
Protocol 1: Implementing a Standardized Metabolite Spike-In Control for Measurement Bias Correction
- Objective: To correct for systematic errors in metabolite quantification during the Test phase, especially in high-throughput screening.
- Materials:
- Procedure:
- Sample Preparation: Add a known, fixed concentration of SIL-IS to each culture sample immediately after quenching metabolism. This controls for losses during sample extraction and preparation.
- Data Acquisition: Acquire data using Data Independent Analysis (DIA) like SWATH-MS for comprehensive metabolite coverage [8].
- Data Normalization: For each metabolite, calculate the ratio of the unlabeled analyte peak area to the SIL-IS peak area. Use this ratio for all subsequent quantitative analyses and model training.
Controlling Experimental Noise
Experimental noise reduces the signal-to-noise ratio in Test data, obscuring genuine cause-and-effect relationships between genetic modifications and phenotypic outcomes. Controlling noise is critical for extracting meaningful insights.
Table 2: Major Sources of Experimental Noise and Control Strategies
| Noise Source | Description | Control Strategy | Expected Outcome |
|---|---|---|---|
| Biological Variability | Stochastic gene expression and cell-to-cell heterogeneity in a clonal population. | Use of well-controlled bioreactors instead of flasks; measure population metrics via flow cytometry. | Reduces variance in measured phenotypes (titer, rate, yield), increasing statistical power. |
| Technical Variability (Sample Processing) | Inconsistencies in sampling, metabolite extraction, and derivatization. | Automation using liquid handling robots; standardized protocols with strict SOPs. | Minimizes non-biological variance, making the data more reproducible. |
| Analytical Instrument Drift | Changes in instrument sensitivity (e.g., LC-MS column degradation) over a long screening run. | Randomized sample run order; inclusion of quality control (QC) reference samples throughout the run. | Prevents systematic time-dependent bias; allows for post-hoc correction of signal drift. |
Experimental Protocol for Noise Reduction
Protocol 2: Automated Microbioreactor Cultivation for High-Throughput Testing
- Objective: To generate highly reproducible phenotype data (growth, productivity) under controlled conditions, minimizing biological and technical noise.
- Materials:
- 48-well or 96-well micro-bioreactor system with online monitoring (OD, pH, DO).
- Automated liquid handling workstation.
- Culture plates with gas-permeable membranes.
- Procedure:
- Inoculation: Use the liquid handler to inoculate from a single, master seed stock into the microbioreactor plates, ensuring uniform starting cell density.
- Cultivation: Run cultures with continuous monitoring and control of temperature and humidity. Use shaking for oxygen transfer.
- Sampling: At defined time points, the liquid handler automatically samples from each well for subsequent end-point analyses (e.g., extracellular metabolites via FIA-HRMS [8]).
- Data Integration: Automatically log all online sensor data and off-line analytical data into a centralized database for the Learn phase.
Strategic Library Design for DBTL Cycles
Library design determines the starting point for the Test phase. A well-designed library efficiently explores the targeted genetic space, providing maximally informative data for the Learn phase.
Library Design Strategies Comparison
Table 3: Library Design Strategies for the Build Phase
| Design Strategy | Principle | Build Method | Best Use Case |
|---|---|---|---|
| Random Mutagenesis | Introduces untargeted mutations across the genome to create diversity. | UV/chemical mutagens; adaptive laboratory evolution (ALE). | Phenotype improvement when genetic basis is unknown; host robustness. |
| Targeted Saturation | Systematically varies all amino acids in a specific enzyme active site. | Multiplex Automated Genome Engineering (MAGE) [8]. | Enzyme engineering for substrate specificity, catalytic efficiency, or themostability [52]. |
| Combinatorial Pathway Assembly | Varies the combination and expression levels of multiple pathway genes. | Golden Gate assembly; Uracil-Specific Excision Reagent (USER) cloning [8]. | Optimizing flux in a heterologous metabolic pathway; balancing expression of multiple genes. |
| Model-Guided Design | Uses genome-scale metabolic models (GSMM) like COBRA to predict beneficial knockouts/overexpressions. | CRISPR-Cas mediated gene knockout/activation; plasmid-based overexpression. | Redirecting central metabolic flux (e.g., in C. glutamicum for L-lysine derived C5 chemicals) [6]. |
Framework for Algorithmic Library Design
The following workflow maps the decision process for selecting and implementing an optimal library design strategy, incorporating modern biofoundry capabilities.
Library Design Strategy Workflow
The Scientist's Toolkit: Research Reagent Solutions
Table 4: Essential Research Reagents for DBTL Cycle Implementation
| Reagent / Tool | Function | Application in DBTL Cycle |
|---|---|---|
| SynBio Open Language (SBOL) | A standardized data format for describing genetic designs [8]. | Design: Ensures reproducible and unambiguous description of genetic parts, devices, and modules for sharing and biofoundry automation. |
| Uracil-Specific Excision Reagent (USER) Cloning | A powerful DNA assembly method [8]. | Build: Facilitates rapid and efficient combinatorial assembly of large metabolic pathway constructs. |
| Multiplex Automated Genome Engineering (MAGE) | A method for introducing multiple targeted mutations simultaneously across the genome [8]. | Build: Enables high-throughput, targeted diversification of genomic sequences (e.g., RBS libraries, promoter swaps). |
| Flow-Injection Analysis HRMS (FIA-HRMS) | A high-throughput method for direct mass spectrometric analysis of culture supernatants without chromatography [8]. | Test: Allows for rapid, quantitative screening of extracellular metabolite levels (e.g., substrates, products, by-products) in thousands of samples. |
| Genome-Scale Metabolic Model (GSMM) | A computational model representing the entire metabolic network of an organism (e.g., C. glutamicum) [8]. | Learn: Used with Constraint-Based Reconstruction and Analysis (COBRA) methods like Flux Balance Analysis (FBA) and Flux Variability Analysis (FVA) to interpret data and generate new testable hypotheses [8]. |
| Elementary Metabolite Units (EMU) | A computational approach for simulating Mass Distribution Vectors (MDVs) in isotopomer analysis [8]. | Learn: Enables the calculation of intracellular metabolic fluxes from 13C-labeling data (MFA), providing critical insights into pathway activity. |
| Ferrous nitrate hexahydrate | Ferrous Nitrate Hexahydrate|Fe(NO₃)₂·6H₂O|CAS 13476-08-9 | |
| Ruthenium hydroxide (Ru(OH)3) | Ruthenium hydroxide (Ru(OH)3), CAS:12135-42-1, MF:H3O3Ru, MW:155.1 g/mol | Chemical Reagent |
In the rigorous framework of Design-Build-Test-Learn (DBTL), a cyclical methodology central to modern systems metabolic engineering, challenges in protein expression and purification are not merely technical setbacks but critical feedback for the next iteration of strain or process design [6]. Achieving high yields of functional protein is a common bottleneck that can stall the development of efficient microbial cell factories. This guide provides a systematic, practical troubleshooting methodology for addressing low protein expression and purification issues, explicitly framed within the DBTL context to help researchers rapidly identify causes, implement corrective actions, and generate actionable knowledge to accelerate the entire engineering cycle.
The DBTL Cycle in Metabolic Engineering
The DBTL cycle is a powerful, iterative framework for optimizing biological systems. In the context of protein production using workhorses like Corynebacterium glutamicum or E. coli, the cycle can be defined as follows [6]:
- Design: Formulating a genetic construct or engineering strategy, such as designing a CRISPR guide RNA or optimizing a codon sequence.
- Build: Executing the design to create a modified biological system, for example, transforming a plasmid or editing the host genome.
- Test: Critically evaluating the output of the built system, which includes measuring protein expression levels, assessing solubility, and purifying the target protein.
- Learn: Analyzing the test data to understand the success or failure of the design, thereby generating insights that inform the next Design phase.
Troubleshooting protein expression and purification is a critical component of the Test and Learn phases. The following workflow diagram (Figure 1) illustrates how a structured troubleshooting process integrates into and reinforces the DBTL cycle, ensuring that each challenge directly contributes to the project's knowledge base.
Troubleshooting Low Protein Expression
Low protein expression can stem from issues at the genetic, cellular, or procedural levels. The following table summarizes the key problem areas, potential causes, and recommended solutions to test during the DBTL cycle.
Table 1: Troubleshooting Guide for Low Protein Expression
| Problem Area | Potential Cause | Recommended Solution & Test Method |
|---|---|---|
| Genetic Design | Suboptimal guide RNA target region (for CRISPR edits) [53] | Design gRNA to target an early exon common to all protein isoforms. Use guide design tools to predict efficacy and minimize off-target effects. |
| Inefficient translation (e.g., rare codons, poor RBS) | Analyze and optimize codon usage for the host. Redesign the Ribosome Binding Site (RBS). Use synthetic genes with host-optimized sequences. | |
| Cellular Health & Selection | Cell line not suitable for CRISPR editing or protein production [53] | Use validated, easy-to-transfect cell lines (e.g., HEK293, HeLa) for initial experiments. For metabolic engineering, select robust microbial hosts like C. glutamicum [6]. |
| Poor cell viability post-transfection/transformation | Check for contamination. Optimize transfection method (electroporation, lipofection) and culture conditions post-transfection. Use a viability stain and cell counter. | |
| Expression Process | Inefficient delivery of CRISPR components [53] | Optimize transfection method (electroporation, lipofection, nucleofection) and the ratio of guide RNA to Cas nuclease for your specific cell line. |
| Suboptimal induction conditions (temperature, timing, inducer concentration) | Perform a time-course and dose-response experiment with the inducer (e.g., IPTG). Test expression at different temperatures (e.g., 30°C vs. 37°C). |
Experimental Protocol: Validating CRISPR Edits
A critical step after attempting a gene knockout is to confirm the edit was successful at both the genomic and protein levels [53].
- Genomic DNA Extraction: Harvest cells and extract genomic DNA using a commercial kit.
- PCR Amplification: Design primers flanking the CRISPR target site and amplify the region.
- Sequence Analysis: Sanger sequence the PCR product and analyze the chromatogram for indels around the cut site. For a mixed population, use TIDE analysis (Tracking of Indels by DEcomposition) to quantify editing efficiency.
- Protein Analysis (Western Blot): Lyse cells and separate proteins via SDS-PAGE. Transfer to a membrane and probe with an antibody against the target protein. A successful knockout should show a complete absence of the protein band.
- Functional Assay: If possible, perform a functional assay to confirm the loss of protein activity.
Troubleshooting Low Protein Yield After Elution
When expression is confirmed but the final purified yield is low, the issue often lies in the purification process itself. The following table outlines common purification pitfalls and how to address them.
Table 2: Troubleshooting Guide for Low Protein Purification Yield
| Problem Area | Potential Cause | Recommended Solution & Test Method |
|---|---|---|
| Lysis & Solubility | Inefficient cell lysis [54] | Increase lysis time; add lysozyme or DNase I; use mechanical methods (sonication, French press). Measure release of total protein via Bradford assay. |
| Target protein in inclusion bodies (insoluble) [54] | Optimize expression conditions (lower temperature, reduce inducer concentration). Use solubility-enhancing tags (e.g., MBP, Trx). Test solubilization with chaotropes (urea, guanidine) and refolding. | |
| Purification | Inadequate binding to resin [54] | Confirm resin compatibility and binding capacity. Increase incubation time. Check that the affinity tag is accessible and not cleaved. Use a binding buffer with appropriate pH and salt. |
| Protein degradation by proteases [54] | Always include a cocktail of protease inhibitors in all buffers. Keep samples on ice or at 4°C throughout the purification. | |
| Elution | Harsh or inefficient elution conditions [54] | For His-tagged proteins, test a gradient or step-elution with imidazole. For other tags, optimize elution buffer pH or use competitive elution. Avoid prolonged incubation in elution buffer. |
Experimental Protocol: Analyzing Purification Yield and Purity
To systematically identify where the protein is being lost, analyze samples from each stage of purification.
- Sample Collection: Collect samples from the following stages: clarified lysate (post-lysis), flow-through (unbound), wash fraction(s), and elution fraction(s).
- SDS-PAGE Analysis: Load equal volumes or normalized protein amounts from each sample onto an SDS-PAGE gel. Include a molecular weight marker.
- Staining and Analysis: Stain the gel with Coomassie Blue or a similar stain. The target protein band should be visible in the lysate, diminish or be absent in the flow-through, and be the dominant band in the elution fractions. The presence of the band in the flow-through indicates poor binding, while its presence in the wash suggests weak washing conditions. Multiple bands in the elution indicate impurity.
- Western Blot (Optional but Recommended): For greater specificity, especially if the target band is faint, perform a Western blot on the same samples using a target-specific antibody.
The following workflow (Figure 2) maps the logical path of the purification troubleshooting process, helping to pinpoint the exact stage of yield loss.
The Scientist's Toolkit: Key Research Reagent Solutions
A successful DBTL cycle relies on high-quality, reliable reagents. The following table details essential materials and their functions in protein expression and purification workflows.
Table 3: Essential Research Reagents for Protein Work
| Reagent / Material | Function & Application in DBTL |
|---|---|
| CRISPR-Cas9 System | Precise genome editing for gene knockout (KO) or knock-in (KI) in the Design and Build phases [53]. Components include guide RNA (synthetic or in vitro transcribed) and Cas nuclease (mRNA or protein). |
| Affinity Chromatography Resins | Core to the Test phase for purifying recombinant proteins. Examples: Ni-NTA for His-tagged proteins, Glutathione Sepharose for GST-tags, Protein A/G for antibodies. |
| Protease Inhibitor Cocktails | Essential for maintaining protein integrity during lysis and purification (Test phase) by preventing degradation by endogenous proteases, thereby protecting yield [54]. |
| Solubility-Enhancing Tags | Genetic fusions (e.g., MBP, Trx, SUMO) used in the Design phase to improve the solubility of poorly expressing proteins, reducing inclusion body formation [54]. |
| Cell Line-Specific Transfection Reagents | Chemical carriers (e.g., lipofection) or electroporation kits optimized for specific cell lines (HEK293, insect cells, etc.) to ensure efficient delivery of genetic material in the Build phase [53]. |
Troubleshooting protein expression and purification is not a linear task but an integral part of the iterative DBTL framework. By systematically testing hypotheses against the potential causes outlined in this guide, researchers can transform failed experiments into valuable learning. The knowledge gained from each troubleshooting cycle—why a particular gRNA design failed, which induction temperature maximizes solubility, or how to optimize an elution gradient—feeds directly and powerfully into the next, more informed Design phase. This rigorous, data-driven approach ensures continuous improvement and accelerates the development of robust microbial cell factories and biopharmaceutical processes.
Benchmarking Success: Validating DBTL Strategies and Emerging Paradigms
In the field of metabolic engineering, the Design-Build-Test-Learn (DBTL) cycle is a foundational framework for the iterative development of microbial cell factories. However, conducting multiple, real-world DBTL cycles is often prohibitively costly and time-consuming, complicating the systematic validation of new methods and strategies [1]. The integration of mechanistic kinetic models offers a powerful solution to this challenge by providing a robust, in silico environment to simulate and optimize DBTL cycles before laboratory implementation. These models use ordinary differential equations (ODEs) derived from the laws of mass action to describe changes in intracellular metabolite concentrations over time, allowing researchers to simulate the effects of genetic perturbations on metabolic flux with biologically relevant interpretations of kinetic parameters [1]. This whitepaper details how mechanistic kinetic models serve as a validation framework for in silico DBTL cycles, enabling more efficient and predictive metabolic engineering.
The Role of Mechanistic Models in the DBTL Cycle
Simulating the Full DBTL Workflow
Mechanistic kinetic models are integrated into each stage of the DBTL cycle to create a predictive, closed-loop system [8]. In the Design phase, models inform the selection of metabolic targets and genetic manipulations. During the Build phase, in silico changes to pathway elements, such as enzyme concentrations (Vmax parameters), are implemented computationally to reflect the assembly of genetic constructs [1]. The Test phase leverages the model to simulate the phenotypic output—such as metabolite fluxes and biomass growth—of these designed strains under simulated bioprocess conditions like batch reactors [1]. Finally, in the Learn phase, the simulated data is used to train machine learning (ML) models, which then recommend improved designs for the next cycle, thus automating and accelerating the strain optimization process [1] [8].
Overcoming Data Scarcity with In Silico Data
A significant advantage of this approach is the generation of rich, synthetic datasets. Publicly available data from multiple, real DBTL cycles are scarce [1]. Mechanistic models overcome this by simulating a vast array of strain designs and their corresponding phenotypes. This data is crucial for training and benchmarking machine learning algorithms—such as gradient boosting and random forest models, which have been shown to perform well in the low-data regime—and for evaluating recommendation systems that guide the next DBTL cycle [1].
Diagram 1: The In Silico DBTL Cycle.
Establishing a Credible Modeling Framework
For in silico DBTL results to be reliable, the computational model must undergo a rigorous credibility assessment [55]. This process, informed by standards like the ASME V&V-40, ensures the model is fit for its specific purpose, or Context of Use (COU).
Key Credibility Activities
- Verification: The process of ensuring the computational model is solved correctly. It answers the question, "Are we solving the equations right?" through checks for numerical accuracy and code correctness [55].
- Validation: The process of determining how well the computational model represents the real-world biological system. It answers, "Are we solving the right equations?" by comparing model predictions with independent experimental data [55].
- Uncertainty Quantification (UQ): Characterizing the impact of uncertainties in model parameters, inputs, and structure on the model's outputs. UQ is essential for understanding the confidence and potential error in model-based predictions [55].
Risk-Informed Credibility Goals
The required level of model credibility is determined through a risk analysis that balances the model influence on a decision with the consequence of an incorrect decision based on that model [55]. A model used for high-impact decisions, such as prioritizing lead candidates for drug development, requires a higher standard of validation than one used for preliminary, internal hypothesis generation.
Diagram 2: Model Credibility Assessment.
A Prototypical Kinetic Model for Pathway Optimization
Model Structure and Integration
To be effective, a kinetic model for DBTL validation must represent a synthetic metabolic pathway embedded within a physiologically relevant host model. A common approach is to integrate a hypothetical pathway (e.g., converting a metabolite A to product G) into an established E. coli core kinetic model [1]. This integrated model simulates:
- Enzyme Kinetics: Each reaction flux is governed by a kinetic mechanism.
- Pathway Topology: The model captures the specific sequence of metabolic conversions.
- Cell Physiology: The pathway is coupled to core metabolism, allowing the simulation of biomass growth and substrate consumption.
- Bioprocess Conditions: The model can be placed in a simulated bioreactor environment, such as a batch culture, to predict product titers over time [1].
Demonstrating the Need for Combinatorial Optimization
Simulations with such integrated models reveal the non-intuitive, non-linear dynamics of metabolic pathways. For example, increasing the concentration of a single enzyme might not increase its reaction flux due to substrate depletion and could even decrease the final product flux [1]. Conversely, the simultaneous, combinatorial optimization of multiple enzyme levels can uncover a global optimum that sequential optimization would miss, thereby validating a core principle of the DBTL approach [1].
Table 1: Core Components of a Kinetic Model for In Silico DBTL
| Model Component | Description | Function in the DBTL Cycle |
|---|---|---|
| Core Host Model | A kinetic model of central metabolism (e.g., E. coli core model) [1] | Provides physiological context and simulates growth and metabolic burden. |
| Integrated Synthetic Pathway | A series of ODEs representing the pathway of interest [1] | Serves as the in silico testbed for designed strain variants. |
| Kinetic Parameters | Vmax, Km, and other constants defining enzyme kinetics [1] | Modified during the "Build" phase to simulate genetic changes (e.g., promoter swaps). |
| Bioprocess Model | Equations simulating a bioreactor (e.g., batch culture) [1] | Allows performance testing under realistic fermentation conditions. |
Quantitative Benchmarking of Machine Learning Methods
The simulated data from kinetic models enables a systematic, quantitative comparison of machine learning (ML) and recommendation algorithms across multiple DBTL cycles—a task difficult to accomplish with experimental data alone [1].
Performance in the Low-Data Regime
Studies using this framework have shown that gradient boosting and random forest models outperform other ML methods when the amount of training data is limited, which is typical of early DBTL cycles. These models have also demonstrated robustness to common experimental challenges like training set biases and measurement noise [1].
Optimizing DBTL Cycle Strategy
The framework allows researchers to test different DBTL strategies. A key finding is that when the total number of strains to be built is limited, an initial cycle with a larger number of strains is more effective for rapid optimization than distributing the same number of strains equally across multiple cycles [1]. This strategy provides a richer initial dataset for the ML models to learn from.
Table 2: Benchmarking ML Performance with a Kinetic Framework
| Evaluation Metric | Description | Key Insight from In Silico DBTL |
|---|---|---|
| Predictive Accuracy | The ability of an ML model to predict strain performance from genetic design. | Gradient boosting and random forest are top performers with limited data [1]. |
| Robustness to Noise | Model performance when simulated experimental noise is added to training data. | The above methods are robust to typical levels of experimental noise [1]. |
| Recommendation Success | The ability of an algorithm to select high-performing strains for the next cycle. | Starting with a larger initial DBTL cycle improves long-term success [1]. |
Experimental Protocol: Implementing an In Silico DBTL Cycle
The following is a detailed methodology for conducting a simulated DBTL cycle using a mechanistic kinetic model.
Protocol: Single-Cycle Simulation and ML Training
Objective: To simulate one complete DBTL cycle for a combinatorial pathway library and train a machine learning model for design recommendation.
Materials & Software:
- A kinetic model implemented in a suitable software environment (e.g., the SKiMpy package in Python) [1].
- A defined DNA library of components (e.g., promoters of varying strengths) for controlling enzyme expression levels.
Procedure:
- Design:
- Define the combinatorial space. For example, specify a set of five distinct expression levels for each of the four enzymes (A, B, C, D) in the pathway, creating a theoretical library of 625 (5^4) unique genetic designs [1].
- Randomly select a subset (e.g., 50 designs) from this full library to constitute the initial DBTL cycle's design pool.
Build:
- For each of the 50 selected designs, programmatically adjust the
Vmaxparameters in the kinetic model to reflect the specified enzyme expression levels [1]. - This step computationally mimics the physical construction of microbial strains.
- For each of the 50 selected designs, programmatically adjust the
Test:
- Run the kinetic model simulation for each of the 50 in silico strains.
- Record the key output metric, typically the product flux (e.g., for metabolite G) at a specific time point or as an integrated value over the simulated fermentation [1].
- This output vector serves as the phenotypic data for the learning phase.
Learn:
- Use the dataset of 50 designs (input features: enzyme levels) and their corresponding simulated product fluxes (output target) to train a supervised machine learning model.
- Benchmark different algorithms (e.g., gradient boosting, random forest, support vector machines) using cross-validation on this data to identify the best performer [1].
- Use the trained model to predict the performance of all remaining untested designs in the full 625-member library.
- Recommend the top N (e.g., 5-10) predicted performers for "construction" in the next simulated DBTL cycle.
Protocol: Multi-Cycle Validation and Strategy Comparison
Objective: To compare the effectiveness of different DBTL cycle strategies over multiple iterations.
Procedure:
- Execute the "Single-Cycle Simulation" protocol as a baseline (Cycle 1).
- For Cycle 2, implement two different strategies in parallel:
- Strategy A (Constant): Build and test a new set of 50 strains.
- Strategy B (Informed): Build and test the top 50 strains recommended by the ML model from Cycle 1.
- Continue both strategies for several cycles, each time using the accumulated data from all previous cycles to retrain the ML model and generate new recommendations.
- Metric for Comparison: Track the highest product flux achieved after each cycle for both strategies. The strategy that reaches a pre-defined performance target in the fewest cycles or with the fewest total strains built is the most efficient.
The Scientist's Toolkit: Essential Research Reagents and Solutions
Table 3: Key Reagents and Computational Tools for Kinetic Modeling and DBTL
| Tool/Reagent | Category | Function/Explanation |
|---|---|---|
| SKiMpy | Software | A Python package for working with symbolic kinetic models; provides the core environment for building and simulating metabolic models [1]. |
| Promoter/RBS Library | DNA Library | A defined set of genetic parts (e.g., promoters, Ribosome Binding Sites) with characterized strengths; provides the sequence space for in silico design variations [1]. |
| ORACLE | Software/Tool | A computational framework for generating and sampling thermodynamically feasible kinetic parameters for metabolic models, enhancing physiological relevance [1]. |
| Gradient Boosting (e.g., XGBoost) | Algorithm | A powerful machine learning algorithm identified via the framework as highly effective for learning from DBTL cycle data in the low-data regime [1]. |
| ASME V&V-40 Standard | Framework | A technical standard providing a formal methodology for assessing the credibility of computational models used in a regulatory or high-stakes context [55]. |
The integration of machine learning (ML) into metabolic engineering has revolutionized the optimization of microbial cell factories through Design-Build-Test-Learn (DBTL) cycles. Among various ML algorithms, Gradient Boosting and Random Forest have demonstrated exceptional performance in guiding combinatorial pathway optimization, particularly in low-data regimes common in early DBTL iterations. This technical analysis examines the benchmark performance of these ensemble methods within DBTL frameworks, providing quantitative comparisons, detailed experimental protocols, and implementation guidelines. Evidence from simulated DBTL cycles demonstrates that these methods outperform other algorithms while maintaining robustness to experimental noise and training set biases, offering metabolic engineers reliable tools for accelerating strain development.
The Design-Build-Test-Learn (DBTL) cycle represents a systematic, iterative framework for engineering biological systems, particularly optimized microbial strains for chemical production. In metabolic engineering, this approach enables rational development of production strains by continually incorporating learning from previous cycles into subsequent designs [6] [3]. The cycle consists of four interconnected phases: (1) Design - selecting genetic modifications or pathway variations based on prior knowledge; (2) Build - implementing these designs through genetic engineering; (3) Test - evaluating strain performance through assays and analytics; and (4) Learn - analyzing results to inform the next design phase [56] [47].
A significant challenge in combinatorial pathway optimization is the combinatorial explosion of possible genetic configurations, making exhaustive experimental testing impractical [56] [57]. Machine learning methods have emerged as powerful tools for the "Learn" phase, enabling researchers to extract meaningful patterns from limited experimental data and propose promising designs for subsequent DBTL cycles [56] [58]. This approach has been successfully applied to optimize diverse products, including p-coumaric acid in yeast [58], dopamine in E. coli [3], and various C5 platform chemicals derived from L-lysine in Corynebacterium glutamicum [6].
Algorithm Fundamentals: Gradient Boosting and Random Forest
Random Forest: Parallel Ensemble Learning
Random Forest operates through bagging (Bootstrap Aggregating), constructing multiple decision trees independently and merging their predictions [59] [60]. Each tree in the forest is trained on a randomly selected subset of the data (bootstrap sampling) and considers only a random subset of features at each split, ensuring diversity among the trees [59]. For classification, the final prediction is determined by majority voting across all trees, while for regression tasks, the average prediction is used [59] [60].
The mathematical representation for a Random Forest can be expressed as:
- Classification:
Final Prediction = mode(Tâ‚(x), Tâ‚‚(x), ..., Tâ‚™(x)) - Regression:
Final Prediction = (1/n) * Σ(Tᵢ(x))
Where Tâ‚, Tâ‚‚, ..., Tâ‚™ represent the individual trees in the forest, x is the input data point, mode gives the most common prediction, and n is the number of trees [59].
Gradient Boosting: Sequential Ensemble Learning
Gradient Boosting builds decision trees sequentially, with each new tree correcting the errors of the previous ensemble [59] [61]. Unlike Random Forest, Gradient Boosting creates an additive model where trees are added one at a time to minimize a loss function. Each new tree is trained on the residual errors (the differences between current predictions and actual values) of the previous model [59] [60] [61].
The algorithm follows this general procedure:
- Initial Prediction: Start with a simple prediction (mean for regression, most frequent class for classification)
- Residual Calculation: Compute residuals between predicted and actual values
- Tree Construction: Train a new tree to predict these residuals
- Model Update: Update predictions by adding the new tree's predictions multiplied by a learning rate
- Iteration: Repeat steps 2-4 until reaching specified number of trees or desired accuracy [59]
Mathematically, the model update at each iteration can be represented as:
F_new(x) = F_old(x) + η * h(x)
Where F(x) represents the current model, h(x) is the new tree trained on residuals, and η is the learning rate controlling the contribution of each tree [59].
Key Algorithmic Differences
Table 1: Fundamental Differences Between Gradient Boosting and Random Forest
| Characteristic | Gradient Boosting | Random Forest |
|---|---|---|
| Model Building | Sequential - trees built one after another | Parallel - trees built independently |
| Bias-Variance | Lower bias, higher variance (more prone to overfitting) | Lower variance, less prone to overfitting |
| Training Approach | Each tree corrects errors of previous trees | Each tree learns from different data subsets |
| Hyperparameter Sensitivity | High sensitivity requiring careful tuning | More robust to suboptimal parameters |
| Computational Complexity | Higher due to sequential training | Lower due to parallel training |
| Robustness to Noise | More sensitive to noisy data and outliers | Generally more robust to noise |
Performance Benchmarks in Metabolic Engineering Applications
Comparative Performance in DBTL Cycles
A 2023 study published in ACS Synthetic Biology provided a mechanistic kinetic model-based framework for consistently comparing machine learning methods across multiple simulated DBTL cycles for combinatorial pathway optimization [56] [57]. This research demonstrated that Gradient Boosting and Random Forest consistently outperformed other machine learning methods, particularly in the low-data regime typical of early DBTL iterations [56].
The study revealed these algorithms maintain robust performance despite common experimental challenges, including training set biases and experimental noise inherent in high-throughput screening data [56] [57]. This robustness is particularly valuable in metabolic engineering applications where measurement variability can significantly impact model performance and subsequent design recommendations.
Large-Scale Algorithm Comparison
Supporting evidence comes from a comprehensive evaluation of 13 machine learning algorithms across 165 classification datasets, which found that Gradient Boosting and Random Forest achieved the lowest average rank (indicating best performance) among all tested methods [62]. The post-hoc analysis underlined the impressive performance of Gradient Boosting, which "significantly outperforms every algorithm except Random Forest at the p < 0.01 level" [62].
Table 2: Performance Characteristics in Metabolic Engineering Context
| Performance Metric | Gradient Boosting | Random Forest |
|---|---|---|
| Low-Data Regime Performance | Excellent | Excellent |
| Robustness to Experimental Noise | Good | Excellent |
| Training Set Bias Robustness | Good | Excellent |
| Handling of Imbalanced Data | Excellent | Good |
| Feature Importance Interpretation | Moderate | Excellent |
| Experimental Resource Optimization | Good | Good |
Experimental Protocols for DBTL Implementation
Framework for Simulated DBTL Cycles
The mechanistic kinetic model-based framework for comparing ML methods in metabolic engineering involves these key methodological steps [56]:
Pathway Representation: Develop kinetic models of metabolic pathways with parameters representing enzyme expression levels, catalytic rates, and metabolite concentrations.
Training Data Generation: Simulate strain variants with different enzyme expression levels and measure resulting metabolic fluxes or product yields.
Model Training: Train ML models (including Gradient Boosting and Random Forest) to predict strain performance from enzyme expression profiles.
Performance Evaluation: Assess model accuracy in predicting metabolic fluxes, particularly with limited training data (simulating early DBTL cycles).
Design Recommendation: Implement algorithms for proposing new strain designs based on model predictions, balancing exploration and exploitation.
Cycle Iteration: Simulate multiple DBTL cycles, using model predictions to select strains for each subsequent "build" phase.
Knowledge-Driven DBTL Cycle for Dopamine Production
A 2025 study demonstrated a knowledge-driven DBTL cycle for optimizing dopamine production in E. coli with these specific experimental components [3]:
In Vitro Pathway Analysis:
- Create crude cell lysate systems expressing pathway enzymes
- Test different relative enzyme expression levels
- Identify optimal expression ratios for maximal dopamine production
In Vivo Strain Construction:
- Engineer high L-tyrosine production host (E. coli FUS4.T2)
- Implement RBS (ribosome binding site) library for fine-tuning expression
- Modulate Shine-Dalgarno sequences while maintaining secondary structure
High-Throughput Screening:
- Cultivate strain variants in minimal medium with appropriate inducers
- Measure dopamine concentrations via HPLC or other analytical methods
- Normalize production by biomass (mg product/g biomass)
Machine Learning Integration:
- Use enzyme expression data and production metrics as training features
- Train models to predict dopamine yield from expression profiles
- Recommend new RBS combinations for subsequent DBTL cycle
Visualization of ML-Guided DBTL Workflows
DBTL Cycle with Machine Learning Integration
DBTL Cycle with Machine Learning Integration
Algorithm Comparison Workflow
Algorithm Comparison Workflow
Research Reagent Solutions for DBTL Experiments
Table 3: Essential Research Reagents and Tools for ML-Guided DBTL Cycles
| Reagent/Tool | Function | Application Example |
|---|---|---|
| RBS Library Variants | Fine-tuning gene expression levels | Modulating enzyme ratios in dopamine pathway [3] |
| Mechanistic Kinetic Models | Simulating metabolic pathway performance | Testing ML methods for combinatorial optimization [56] |
| Cell-Free Protein Synthesis Systems | Testing enzyme expression without cellular constraints | In vitro pathway optimization before in vivo implementation [3] |
| Automated Recommendation Algorithms | Proposing new strain designs based on ML predictions | Balancing exploration and exploitation in design selection [56] [58] |
| Biosensor-Enabled Screening | High-throughput measurement of metabolic fluxes | Generating training data for ML models [58] |
Implementation Guidelines for Metabolic Engineers
When to Select Each Algorithm
Based on the benchmark performance in metabolic engineering applications:
Choose Random Forest when:
- Working with noisy experimental data or potential outliers
- Requiring faster training times and computational efficiency
- Needing interpretable feature importance to guide biological insight
- Building initial models with limited hyperparameter tuning resources
- Operating in lower-data regimes where overfitting is a concern [59] [56] [60]
Choose Gradient Boosting when:
- Predictive accuracy is the primary concern
- Working with relatively clean, well-characterized datasets
- Having sufficient computational resources for extensive hyperparameter tuning
- Dealing with imbalanced data where certain high-performance variants are rare
- Addressing problems with complex, non-linear relationships between pathway modifications and production [59] [62]
Hyperparameter Optimization Guidelines
For Gradient Boosting in metabolic engineering applications:
- Learning rate: Typically between 0.01-0.2, with lower values requiring more trees
- Number of trees: Balance between performance and overfitting (typically 100-500)
- Tree depth: Shallow trees (3-8 levels) often perform well as weak learners
- Subsampling: Use less than 1.0 to reduce overfitting [59] [61]
For Random Forest in metabolic engineering applications:
- Number of trees: Typically 100-500, with diminishing returns beyond certain point
- Features per split: Typically √p or log₂(p) where p is total features
- Tree depth: Can allow full depth without pruning due to averaging effect
- Bootstrap sample size: Typically 60-80% of total data [59] [60]
Gradient Boosting and Random Forest have established themselves as benchmark machine learning methods for guiding DBTL cycles in metabolic engineering. Their demonstrated performance in low-data regimes, robustness to experimental noise, and ability to extract meaningful patterns from complex biological data make them invaluable tools for accelerating strain development. As automated biofoundries and high-throughput screening technologies continue to generate larger experimental datasets, the strategic implementation of these ensemble methods will play an increasingly critical role in optimizing microbial cell factories for sustainable chemical production.
The Design-Build-Test-Learn (DBTL) cycle has long served as the fundamental framework for systematic engineering in synthetic biology and metabolic engineering. This iterative process streamlines efforts to build biological systems by providing a structured approach to engineering until desired functions are achieved [5]. However, recent advancements in machine learning (ML) and high-throughput testing platforms are driving a potential paradigm shift. A new framework, known as LDBT (Learn-Design-Build-Test), repositions machine learning at the forefront of the biological engineering cycle [5] [33]. This comparative analysis examines the technical foundations, implementation protocols, and practical applications of both paradigms, providing researchers with a comprehensive guide to their distinctive strengths and appropriate use cases within metabolic engineering research.
Core Conceptual Frameworks and Workflows
The Traditional DBTL Cycle
The traditional DBTL cycle is characterized by a linear, iterative workflow where each phase sequentially informs the next. In the Design phase, researchers define objectives and design biological parts or systems using domain knowledge and computational modeling. The Build phase involves synthesizing DNA constructs and introducing them into characterization systems (e.g., bacterial chassis). The Test phase experimentally measures the performance of engineered biological constructs. Finally, the Learn phase analyzes collected data to inform the next design round, creating a loop of continuous improvement [5] [63]. This framework has proven effective in streamlining biological engineering efforts, with cycle automation becoming increasingly sophisticated through biofoundries [10] [64].
The Machine-Learning-First LDBT Paradigm
The emerging LDBT paradigm fundamentally reorders the workflow, starting with a Learning phase powered by machine learning models that interpret existing biological data to predict meaningful design parameters [5] [33]. This learning-first approach enables researchers to refine design hypotheses before constructing biological parts, potentially circumventing costly trial-and-error. The subsequent Design phase leverages computational predictions to create optimized biological designs, which are then Built and Tested using rapid, high-throughput platforms like cell-free transcription-translation systems [33]. This reordering aims to leverage the predictive power of machine learning to reduce experimental iterations and accelerate convergence on functional solutions.
Table 1: Core Workflow Comparison Between DBTL and LDBT Cycles
| Phase | Traditional DBTL Approach | Machine-Learning-First LDBT Approach |
|---|---|---|
| Starting Point | Design based on domain knowledge and objectives | Learning from existing biological data using ML models |
| Primary Driver | Empirical iteration and sequential improvement | Predictive modeling and zero-shot design |
| Build Emphasis | In vivo systems in living chassis | Cell-free systems for rapid prototyping |
| Testing Methodology | Cellular assays and functional measurements | High-throughput cell-free screening |
| Learning Mechanism | Post-hoc analysis of experimental results | Continuous model improvement with new data |
| Cycle Objective | Iterative refinement through multiple cycles | Single-cycle convergence where possible |
Visualizing the Workflow Differences
The following diagram illustrates the fundamental structural differences between the traditional DBTL cycle and the machine-learning-first LDBT paradigm:
Technical Implementation and Methodological Approaches
Machine Learning Tools and Algorithms
The LDBT paradigm leverages sophisticated machine learning approaches that fall into several key categories:
Protein-Focused Models: Sequence-based protein language models such as ESM and ProGen are trained on evolutionary relationships between protein sequences and can predict beneficial mutations and infer protein functions [5]. Structure-based models like MutCompute and ProteinMPNN use deep neural networks trained on protein structures to associate amino acids with their chemical environment, enabling prediction of stabilizing substitutions [5]. These have demonstrated success in engineering hydrolases for PET depolymerization with improved stability and activity.
Function-Optimization Models: Tools like Prethermut and Stability Oracle predict effects of mutations on thermodynamic stability using machine learning trained on experimental data [5]. DeepSol predicts protein solubility from primary sequences, representing efforts to predict functional characteristics directly [5].
Recommendation Systems: The Automated Recommendation Tool (ART) combines machine learning with probabilistic modeling to recommend strain designs for subsequent engineering cycles [65]. This tool uses an ensemble approach adapted to synthetic biology's specific needs, including small dataset sizes and uncertainty quantification [65].
Table 2: Key Machine Learning Tools and Their Applications in the LDBT Paradigm
| Tool Name | ML Approach | Primary Application | Validated Use Case |
|---|---|---|---|
| ESM | Protein language model | Zero-shot prediction of protein functions | Predicting beneficial mutations and solvent-exposed amino acids [5] |
| ProteinMPNN | Structure-based deep learning | Protein sequence design given backbone structure | TEV protease engineering with improved catalytic activity [5] |
| MutCompute | Deep neural network | Residue-level optimization based on local environment | Engineering PET depolymerization hydrolases [5] |
| Stability Oracle | Graph-transformer architecture | Predicting thermodynamic stability changes (ΔΔG) | Identifying stabilizing mutations [5] |
| Automated Recommendation Tool (ART) | Bayesian ensemble methods | Recommending strain designs for next DBTL cycle | Optimizing tryptophan production in yeast [65] |
| DeepSol | Deep learning on k-mers | Predicting protein solubility from sequence | Solubility optimization for recombinant proteins [5] |
Experimental Platforms and High-Throughput Methodologies
Cell-Free Transcription-Translation Systems play a pivotal role in the LDBT paradigm by enabling rapid testing phases. These systems leverage protein biosynthesis machinery from cell lysates or purified components to activate in vitro transcription and translation [5]. Their advantages include:
- Speed: Protein production exceeding 1 g/L in under 4 hours [5]
- Scalability: Reactions scalable from picoliter to kiloliter scales [5]
- Tolerance: Production of products toxic to living cells [5]
- Flexibility: Customizable reaction environments and incorporation of non-canonical amino acids [5]
Automation and Biofoundries enhance both paradigms but are particularly crucial for LDBT implementation. Automated biofoundries integrate laboratory automation with data management systems, enabling high-throughput construction and testing [10] [9]. The iPROBE platform exemplifies this approach, using cell-free systems and neural networks to optimize biosynthetic pathways, resulting in 20-fold improvement of 3-HB production in Clostridium [5].
Detailed Experimental Protocol: Implementing an Automated DBTL Pipeline
For researchers implementing automated DBTL cycles, the following protocol outlines key steps based on successful implementations:
Design Phase Protocol:
- Pathway Selection: Use computational tools like RetroPath for automated pathway selection from target compounds [10]
- Enzyme Selection: Employ tools like Selenzyme for selecting optimal enzymes for each pathway step [10]
- DNA Part Design: Utilize software such as PartsGenie for designing reusable DNA parts with optimized ribosome-binding sites and codon optimization [10]
- Library Design: Create combinatorial libraries of pathway designs, then apply Design of Experiments (DoE) to reduce library size while maintaining representative diversity [10]
Build Phase Protocol:
- Automated DNA Assembly: Use ligase cycling reaction or Golden Gate assembly on robotic liquid handling platforms [10] [9]
- Quality Control: Implement high-throughput plasmid purification, restriction digest analysis, and sequence verification [10]
- Chassis Preparation: Transform constructs into production chassis using standardized protocols
Test Phase Protocol:
- Cultivation: Employ automated 96-deepwell plate growth and induction protocols [10]
- Metabolite Extraction: Implement automated extraction protocols for target compounds and intermediates
- Analytical Measurement: Utilize fast UPLC coupled with tandem mass spectrometry for quantitative screening [10]
- Data Processing: Apply custom scripts for data extraction and processing [10]
Learn Phase Protocol:
- Statistical Analysis: Identify relationships between production levels and design factors using statistical methods [10]
- Machine Learning: Train models on collected data to predict performance of new designs [65]
- Recommendation Generation: Use tools like ART to propose designs for subsequent cycles [65]
Case Studies and Performance Comparison
Traditional DBTL Success: Dopamine Production in E. coli
A 2025 study demonstrated the successful application of a knowledge-driven DBTL cycle to optimize dopamine production in Escherichia coli [3]. Researchers employed a rational strain engineering approach with the following key steps:
- Design: Targeted engineering of E. coli genome to increase L-tyrosine production as dopamine precursor
- Build: Construction of strains with heterologous expression of HpaBC and Ddc genes
- Test: High-throughput screening of dopamine production using analytical methods
- Learn: Analysis of rate-limiting steps to inform subsequent design iterations
This approach achieved dopamine production of 69.03 ± 1.2 mg/L, representing a 2.6 to 6.6-fold improvement over previous state-of-the-art production [3]. The success highlights how traditional DBTL remains powerful when guided by mechanistic understanding and hypothesis-driven design.
LDBT Implementation: Flavonoid Pathway Optimization
The application of an automated DBTL pipeline to flavonoid production showcases the integration of machine learning and high-throughput methods [10]. In this implementation:
- Initial library design created 2592 possible pathway configurations
- Design of Experiments reduced this to 16 representative constructs
- Statistical analysis identified vector copy number as the strongest factor affecting production
- A second design cycle incorporating these insights improved pinocembrin production by 500-fold, achieving titers up to 88 mg Lâ»Â¹ [10]
This case demonstrates how even partial automation and data-driven learning can dramatically accelerate pathway optimization.
Performance Benchmarking and Quantitative Comparisons
Table 3: Quantitative Comparison of DBTL vs. LDBT Performance Metrics
| Performance Metric | Traditional DBTL Approach | Machine-Learning-First LDBT |
|---|---|---|
| Cycle Duration | Weeks to months per cycle [10] | Days to weeks with cell-free systems [5] [33] |
| Experimental Throughput | Limited by cellular growth and cloning | 100,000+ reactions using droplet microfluidics [5] |
| Data Generation Capacity | Moderate, constrained by cellular assays | Megascale data generation capabilities [5] |
| Success Rate (First Cycle) | Low, requires multiple iterations | Improved through zero-shot predictions [5] |
| Resource Requirements | High per strain built and tested | Focused on promising designs, reducing waste [33] |
| Optimization Efficiency | 2.6-6.6 fold improvement in demonstrated cases [3] | 500-fold improvement in demonstrated cases [10] |
The Scientist's Toolkit: Essential Research Reagents and Platforms
Implementing either DBTL or LDBT approaches requires specific research tools and platforms. The following table details essential components for establishing these engineering cycles in research settings:
Table 4: Essential Research Reagent Solutions for DBTL and LDBT Implementation
| Tool Category | Specific Tools/Platforms | Function | Compatible Paradigm |
|---|---|---|---|
| DNA Design Software | PartsGenie, PlasmidGenie, TeselaGen | Automated DNA part design and assembly protocol generation | Both (Essential for LDBT) [10] [9] |
| Machine Learning Tools | ART, ESM, ProteinMPNN, Stability Oracle | Predictive modeling and design recommendation | Primarily LDBT [5] [65] |
| Cell-Free Systems | TX-TL systems, crude cell lysates | Rapid in vitro protein expression and testing | Primarily LDBT [5] [33] |
| Automated Liquid Handlers | Tecan, Beckman Coulter, Hamilton Robotics | High-precision liquid handling for assembly and screening | Both (Essential for scale) [9] |
| Analytical Instruments | UPLC-MS/MS, Plate Readers, NGS platforms | Quantitative measurement of products and intermediates | Both [10] [9] |
| Data Management Platforms | JBEI-ICE, TeselaGen Platform | Centralized data storage, sample tracking, and analysis | Both (Essential for LDBT) [10] [9] |
| DNA Synthesis Providers | Twist Bioscience, IDT, GenScript | High-quality DNA fragment and gene synthesis | Both [9] |
Integration Pathways and Future Outlook
The relationship between DBTL and LDBT is not strictly sequential but rather represents complementary approaches. A hybrid framework that incorporates elements of both may represent the most pragmatic path forward:
This integrated approach leverages the predictive power of machine learning while maintaining the physiological relevance of traditional in vivo validation. As machine learning models continue to improve and more comprehensive training datasets become available, the balance may shift further toward LDBT approaches. However, the complex cellular context of metabolic engineering ensures that traditional DBTL cycles will remain relevant for the foreseeable future, particularly for fine-tuning pathway performance in industrial production strains and addressing complex regulatory challenges that exceed current predictive capabilities.
The comparative analysis reveals that both traditional DBTL and machine-learning-first LDBT paradigms offer distinct advantages for metabolic engineering research. The traditional DBTL cycle provides a robust, reliable framework for hypothesis-driven engineering with proven success across numerous applications. Its strength lies in accommodating biological complexity and providing mechanistic insights through iterative experimentation. In contrast, the LDBT paradigm offers accelerated design cycles and reduced experimental burden through predictive modeling and rapid prototyping, excelling in exploration of large design spaces and data-rich scenarios.
For research teams, the optimal approach depends on specific project goals, available resources, and existing knowledge about the target system. Traditional DBTL remains advantageous for novel pathway engineering with limited prior data, while LDBT shows strong promise for optimizing characterized systems and exploring complex design spaces. As synthetic biology continues its trajectory toward greater predictability and efficiency, the integration of both approaches within a flexible, automated infrastructure will likely drive the next generation of advances in metabolic engineering and pharmaceutical development.
The Design-Build-Test-Learn (DBTL) cycle has long been a cornerstone of engineering disciplines, and its application in metabolic engineering is revolutionizing the development of microbial cell factories. This iterative process, which involves designing genetic modifications, building DNA constructs, testing the resulting strains, and learning from the data to inform the next cycle, is fundamental for optimizing the production of biofuels, pharmaceuticals, and fine chemicals. However, traditional DBTL cycles are often hampered by their slow pace, high costs, and reliance on researcher intuition, creating bottlenecks in bioprocess development. The integration of advanced automation and artificial intelligence (AI) is now transforming this workflow, leading to unprecedented gains in both economic efficiency and development speed. This whitepaper assesses these gains through quantitative data, detailed experimental protocols, and visualizations, providing researchers and drug development professionals with a clear understanding of how modern technologies are accelerating metabolic engineering.
The Evolution of the DBTL Cycle in Metabolic Engineering
The classical DBTL cycle involves sequential, often manual, steps. The Design phase relies on prior knowledge and homology to select enzymes and pathways. The Build phase involves molecular biology techniques to assemble genetic constructs. The Test phase cultivates engineered strains and measures product titers, rates, and yields (TRYs). Finally, the Learn phase uses statistical analysis to identify successful designs for the next iteration. A significant limitation of this approach is its low throughput and the high human resource cost at each stage, leading to long development timelines.
The contemporary DBTL cycle, in contrast, is characterized by the integration of automation, robotics, and AI. This transformation creates a high-throughput, closed-loop system where AI models use experimental data to autonomously design improved variants for the next round of testing. This shift addresses key bottlenecks:
- Automation and Robotics: Automated liquid handlers, robotic arms, and integrated biofoundries like the Illinois Biological Foundry for Advanced Biomanufacturing (iBioFAB) enable 24/7 operation for the Build and Test phases, drastically increasing throughput and reproducibility while reducing human labor [66].
- Artificial Intelligence and Machine Learning: AI, particularly machine learning (ML) and large language models (LLMs), has revolutionized the Design and Learn phases. These tools can navigate vast biological design spaces more efficiently than human intuition, identifying non-obvious solutions and optimizing pathways based on predictive models [66] [67].
Table 1: Core Components of a Modern, Automated DBTL Framework
| DBTL Phase | Traditional Approach | Modern Automated/AI Approach | Key Enabling Technologies |
|---|---|---|---|
| Design | Manual literature review, homology-based enzyme selection | AI-powered enzyme and pathway selection; predictive modeling using LLMs | RetroPath, Selenzyme, Protein LLMs (e.g., ESM-2), Ensemble Modeling [10] [66] [67] |
| Build | Manual cloning, site-directed mutagenesis | Automated DNA assembly, high-fidelity robotic cloning | iBioFAB, Ligase Cycling Reaction (LCR), Automated PCR setup and purification [10] [66] |
| Test | Shake-flask cultures, manual sampling and extraction | High-throughput cultivation in microtiter plates, automated analytics | Robotic liquid handling, integrated UPLC-MS/MS, online sensors [10] |
| Learn | Basic statistical analysis (e.g., ANOVA) | Machine learning model training for predictive fitness assessment | Bayesian Optimization, Low-N ML models, Statistical DoE analysis [10] [66] |
Quantitative Assessment of Efficiency Gains
The implementation of automated and AI-driven DBTL cycles has yielded demonstrable and significant improvements in both the time required for engineering campaigns and the functional outcomes of the engineered systems.
Temporal Efficiency and Throughput
A landmark study demonstrated an AI-powered autonomous platform that engineered two enzymes for dramatically improved activity within just four weeks [66]. This platform required the construction and characterization of fewer than 500 variants for each enzyme to achieve its goals, showcasing highly efficient navigation of sequence space. Another study on an automated DBTL pipeline for fine chemical production established a production pathway improved by 500-fold in just two DBTL cycles [10]. These examples highlight a compression of development timelines from years or months to weeks.
Performance and Economic Output
The performance gains from AI-driven engineering are substantial. In enzyme engineering, studies have reported:
- A 90-fold improvement in substrate preference and a 16-fold improvement in ethyltransferase activity for Arabidopsis thaliana halide methyltransferase (AtHMT) [66].
- A 26-fold improvement in activity at neutral pH for Yersinia mollaretii phytase (YmPhytase) [66].
In broader metabolic engineering for biofuels, advances include a 91% biodiesel conversion efficiency from lipids and a 3-fold increase in butanol yield in engineered Clostridium spp. [68]. These enhanced performance metrics directly translate to improved economic viability by increasing the volumetric productivity and yield of the bioprocess, reducing the cost per unit of product.
Table 2: Measured Economic and Temporal Gains from Automated DBTL Implementations
| Application Area | Key Performance Indicator | Result with Automated/AI DBTL | Citation |
|---|---|---|---|
| Enzyme Engineering | Campaign Duration | 4 weeks for 16-90 fold activity improvement | [66] |
| Enzyme Engineering | Screening Efficiency | <500 variants built and characterized per enzyme | [66] |
| Fine Chemical Production | Improvement in Titer (Pinocembrin) | 500-fold increase in 2 DBTL cycles | [10] |
| Biofuel Production | Butanol Yield in Clostridium | 3-fold increase | [68] |
| Biofuel Production | Biodiesel Conversion Efficiency | 91% from lipids | [68] |
Experimental Protocols for an Automated DBTL Workflow
The following detailed methodology is adapted from next-generation biofoundry operations, illustrating how an automated DBTL cycle is executed for a protein engineering campaign.
Protocol: Autonomous Enzyme Engineering on a Biofoundry
Objective: To improve a specific enzymatic property (e.g., activity, specificity, stability) through iterative, AI-driven cycles. Strain/Materials: E. coli or yeast as an expression chassis; wild-type gene of the target enzyme.
Modular Workflow Steps:
Module 1: AI-Driven Library Design
- Input: The wild-type amino acid sequence of the target enzyme.
- Process: A protein Large Language Model (LLM) like ESM-2 and an epistasis model (e.g., EVmutation) are used to generate a list of candidate single-point mutations. The models predict which amino acid substitutions are most likely to improve the desired function, maximizing the diversity and quality of the initial variant library [66].
- Output: A list of ~150-200 gene variants to be synthesized.
Module 2: Automated DNA Construction
- Process: The biofoundry executes a high-fidelity assembly-based mutagenesis method.
- Automated Mutagenesis PCR: Robotic platforms set up PCR reactions to introduce mutations.
- DpnI Digestion & Purification: Automated digestion of template DNA and purification of PCR products.
- HiFi DNA Assembly: Robotic assembly of the mutated gene fragments into a plasmid backbone.
- Transformation: Automated transformation of the assembly reaction into E. coli cells in a 96-well format.
- Colony Picking & Plasmid Purification: A robotic arm picks individual colonies into deep-well plates for culturing, followed by automated plasmid DNA purification [66].
- Quality Control: A subset of constructs is randomly sequenced to verify accuracy (>95% success rate).
- Process: The biofoundry executes a high-fidelity assembly-based mutagenesis method.
Module 3: High-Throughput Screening & Characterization
- Process: The biofoundry conducts expression and functional assays.
- Protein Expression: Automated induction of protein expression in 96-deepwell plates.
- Cell Lysis: Robotic harvest and lysis of cells.
- Enzyme Assay: The assay (e.g., a colorimetric or fluorometric readout) is performed robotically by mixing the cell lysate with substrate in a plate reader. The platform is programmed to measure the initial rate of reaction for each variant [66].
- Data Capture: Absorbance or fluorescence data is automatically logged into a central database.
- Process: The biofoundry conducts expression and functional assays.
Module 4: Machine Learning and Next-Cycle Design
- Process: The functional data from the tested variants is used to train a machine learning model (e.g., a Bayesian optimizer or a low-N ML model). This model learns the complex relationship between sequence changes and functional output.
- Output: The trained model predicts a new set of variants, often combining beneficial mutations from the first round, expected to show further improvement. This list is autonomously fed back into Module 1, closing the DBTL loop [66].
Diagram 1: Automated DBTL cycle for enzyme engineering.
The Scientist's Toolkit: Essential Research Reagents and Solutions
Implementing an automated DBTL cycle requires a suite of specialized reagents, software, and hardware. The following table details key components essential for setting up such a pipeline.
Table 3: Research Reagent Solutions for an Automated DBTL Pipeline
| Item Name | Function / Application | Specification / Notes |
|---|---|---|
| iBioFAB (Illinois Biological Foundry) | Integrated robotic platform for end-to-end automation of biological experiments. | Enables modular, continuous workflows for DNA construction, transformation, and screening [66]. |
| Ligase Cycling Reaction (LCR) Reagents | For automated, highly efficient assembly of combinatorial DNA libraries. | Preferred over traditional methods for its robustness and suitability for robotic setup [10]. |
| ESM-2 (Evolutionary Scale Model) | Protein Large Language Model for in silico variant design and fitness prediction. | A transformer model trained on global protein sequences to predict the likelihood of beneficial mutations [66]. |
| Flux Balance Analysis (FBA) Software | Constraint-based modeling for predicting metabolic flux distributions in genome-scale models. | Used to identify gene knockout or overexpression targets to optimize metabolic pathways [67]. |
| HiFi DNA Assembly Master Mix | Enzyme mix for accurate assembly of multiple DNA fragments. | Critical for the automated Build phase to ensure high-fidelity construction of variant libraries [66]. |
| UPLC-MS/MS Systems | For automated, high-throughput quantification of target metabolites and pathway intermediates. | Provides rapid and sensitive data for the Test phase, essential for generating high-quality training data for ML [10]. |
The integration of automation and artificial intelligence into the DBTL cycle represents a paradigm shift for metabolic engineering and drug development. The quantitative evidence is clear: these technologies deliver order-of-magnitude improvements in engineering efficiency, compressing development timelines from years to weeks and dramatically enhancing the performance of biocatalysts and microbial strains. As these platforms become more generalized and accessible, they promise to accelerate the creation of sustainable bioprocesses for chemical, fuel, and pharmaceutical production. For researchers and organizations, investing in the infrastructure and expertise to leverage these automated DBTL cycles is no longer a frontier advantage but a necessity to remain competitive in the rapidly evolving landscape of biotechnology.
Conclusion
The DBTL cycle has firmly established itself as an indispensable, iterative engine for advancing metabolic engineering. The integration of automation, high-throughput analytics, and particularly machine learning is transforming DBTL from a largely empirical process toward a more predictive and efficient discipline. The emergence of paradigms like LDBT, which places Learning first, underscores a pivotal shift. Future directions point toward closed-loop, self-optimizing biofoundries and the application of foundational AI models trained on megascale biological data. For biomedical and clinical research, these accelerated and knowledge-driven DBTL workflows promise to drastically shorten development timelines for microbial production of complex therapeutics, diagnostic agents, and valuable fine chemicals, ultimately enabling more rapid translation from lab-scale discovery to clinical and industrial impact.
References
- 1. Simulated Design–Build–Test–Learn Cycles for Consistent ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10510747/]
- 2. Analytics for Metabolic Engineering - PMC - PubMed Central [https://pmc.ncbi.nlm.nih.gov/articles/PMC4561385/]
- 3. The knowledge driven DBTL cycle provides mechanistic ... [https://microbialcellfactories.biomedcentral.com/articles/10.1186/s12934-025-02729-6]
- 4. Engineering - Iterative DBTL Cycles | LYON - iGEM 2025 [https://2025.igem.wiki/lyon/engineering]
- 5. combining machine learning and rapid cell-free testing [https://www.nature.com/articles/s41467-025-65281-2]
- 6. Recent advances in the Design-Build-Test-Learn (DBTL ... [https://pubmed.ncbi.nlm.nih.gov/40195836/]
- 7. Learning the way toward high-precision biological design [https://pmc.ncbi.nlm.nih.gov/articles/PMC10231942/]
- 8. Automating the design-build-test-learn cycle towards next- ... [https://www.sciencedirect.com/science/article/pii/S187167842300002X]
- 9. DBTL Cycle Advancement: Software Elevates Biotech R&D [https://teselagen.com/blog/revolutionizing-rd-dbtl-cycle/]
- 10. An automated Design-Build-Test-Learn pipeline for ... [https://www.nature.com/articles/s42003-018-0076-9]
- 11. Approaches to Debottlenecking and Process Optimization [https://www.bioprocessintl.com/chromatography/approaches-to-debottlenecking-and-process-optimization]
- 12. Application of combinatorial optimization strategies in ... [https://www.nature.com/articles/s41467-020-16175-y]
- 13. Combinatorial pathway optimization for streamlined ... [https://www.sciencedirect.com/science/article/abs/pii/S0958166917300472]
- 14. Pathway Evolution Through a Bottlenecking ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11005738/]
- 15. Combinatorial vs Sequential Optimization for Metabolic ... [https://www.genscript.com/synthetic-biology-news/combinatorial-vs-sequential-optimization-for-metabolic-pathway-engineering.html]
- 16. Combinatorial metabolic pathway assembly approaches ... [https://www.sciencedirect.com/science/article/abs/pii/S1096717620301853]
- 17. Metabolic engineering: Tools and applications - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC11610952/]
- 18. a solution to involution of design–build–test–learn cycle [https://www.sciencedirect.com/science/article/abs/pii/S0958166922000453]
- 19. BIOS: a bio-intelligent DBTL approach for sustainable bio- ... [https://www.wur.nl/en/project/bios-a-bio-intelligent-dbtl-approach-for-sustainable-bio-manufacturing.htm]
- 20. The bio-intelligent DBTL cycle, a key enabler catalysing the ... [https://cris.vtt.fi/en/projects/the-bio-intelligent-dbtl-cycle-a-key-enabler-catalysing-the-indus]
- 21. An automated Design-Build-Test-Learn pipeline for ... [https://pubmed.ncbi.nlm.nih.gov/30271948/]
- 22. Engineering an Escherichia coli strain for enhanced ... [https://microbialcellfactories.biomedcentral.com/articles/10.1186/s12934-024-02582-z]
- 23. Metabolic engineering of Escherichia coli for (2S) [https://pubmed.ncbi.nlm.nih.gov/23246524/]
- 24. Metabolic engineering of Escherichia coli for (2S) [https://www.sciencedirect.com/science/article/abs/pii/S1096717612001279]
- 25. Enhanced pinocembrin production in Escherichia coli by ... [https://www.nature.com/articles/srep32640]
- 26. The knowledge driven DBTL cycle provides mechanistic ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12084978/]
- 27. The knowledge driven DBTL cycle provides mechanistic ... [https://pubmed.ncbi.nlm.nih.gov/40380156/]
- 28. Rationally reduced libraries for combinatorial pathway ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4821882/]
- 29. Combinatorial metabolic engineering using an orthogonal ... [https://www.nature.com/articles/s41467-017-01695-x]
- 30. Targeted mutagenesis and high - throughput screening of diversified... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10400447/]
- 31. Multiplex base editing of RBSs rewires Bacillus subtilis ... [https://www.sciencedirect.com/science/article/abs/pii/S1096717625001594]
- 32. A Step-by-Step Protocol for COMPASS, a Synthetic Biology ... [https://pubmed.ncbi.nlm.nih.gov/32809205/]
- 33. LDBT: Machine Learning Meets Rapid Cell-Free Testing [https://bioengineer.org/ldbt-machine-learning-meets-rapid-cell-free-testing/]
- 34. An AI-driven workflow for the accelerated optimization of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12529354/]
- 35. Color Patterns Every Developer Needs to Understand ... [https://medium.com/@rameshkannanyt0078/color-patterns-every-developer-needs-to-understand-2025-edition-56b423a1e411]
- 36. Color contrast - Accessibility - MDN Web Docs - Mozilla [https://developer.mozilla.org/en-US/docs/Web/Accessibility/Guides/Understanding_WCAG/Perceivable/Color_contrast]
- 37. Graphic design color palettes that stick [2025 edition] [https://www.todaymade.com/blog/graphic-design-color-palettes]
- 38. Yellow, Purple, and the Myth of “Accessibility Limits Color ... [https://stephaniewalter.design/blog/yellow-purple-and-the-myth-of-accessibility-limits-color-palettes/]
- 39. Cell-Free Systems to Mimic and Expand Metabolism - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC11852204/]
- 40. Frontiers in biofoundry: opportunities and challenges [https://www.frontiersin.org/journals/synthetic-biology/articles/10.3389/fsybi.2025.1630026/full]
- 41. Common principles and best practices for engineering ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8323346/]
- 42. Getting the Right Clones in an Automated Manner [https://www.mdpi.com/2306-5354/11/9/892]
- 43. DNA Synthesis Market Size, Share | Growth Report [2025- ... [https://www.fortunebusinessinsights.com/dna-synthesis-market-109799]
- 44. The Rise of Commercial Gene Synthesis [https://synbio-tech.com/the-evolution-of-commercial-gene-synthesis-history-of-trends-ai-innovations]
- 45. Made-to-order DNA goes big: new tech doubles size of ... [https://www.nature.com/articles/d41586-025-02261-y]
- 46. Automated high-throughput DNA synthesis and assembly [https://www.sciencedirect.com/science/article/pii/S2405844024029980]
- 47. The Design-Build-Test-Learn cycle for metabolic ... [https://orbit.dtu.dk/en/publications/the-design-build-test-learn-cycle-for-metabolic-engineering-of-st/]
- 48. A machine learning Automated Recommendation Tool for ... [https://www.nature.com/articles/s41467-020-18008-4]
- 49. Questions, data and models underpinning metabolic ... [https://www.frontiersin.org/journals/systems-biology/articles/10.3389/fsysb.2022.998048/full]
- 50. Combining mechanistic and machine learning models for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7519671/]
- 51. Machine and deep learning meet genome-scale metabolic ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6622478/]
- 52. Improving metabolic engineering design with enzyme ... [https://www.sciencedirect.com/science/article/pii/S1096717625000850]
- 53. Irregular Protein Expression After CRISPR Edits [https://www.synthego.com/blog/crispr-irregular-protein-expression/]
- 54. How to Troubleshoot Low Protein Yield After Elution [https://synapse.patsnap.com/article/how-to-troubleshoot-low-protein-yield-after-elution]
- 55. In silico trials: Verification, validation and uncertainty ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7883933/]
- 56. Simulated Design-Build-Test-Learn Cycles for Consistent ... [https://research.tudelft.nl/en/publications/simulated-design-build-test-learn-cycles-for-consistent-compariso/]
- 57. Simulated Design-Build-Test-Learn Cycles for Consistent ... [https://www.broadinstitute.org/publications/broad1340736]
- 58. Machine Learning-Assisted Pathway Optimization in Large ... [https://www.semanticscholar.org/paper/Machine-Learning-Assisted-Pathway-Optimization-in-a-Lent-Hoek/57c8ce839b48891d4155bfdd420af75db16b3362]
- 59. Gradient Boosting vs. Random Forest: Which Ensemble ... [https://medium.com/@hassaanidrees7/gradient-boosting-vs-random-forest-which-ensemble-method-should-you-use-9f2ee294d9c6]
- 60. Gradient Boosting vs Random Forest [https://www.geeksforgeeks.org/machine-learning/gradient-boosting-vs-random-forest/]
- 61. An Introduction to Gradient Boosting Decision Trees [https://www.machinelearningplus.com/machine-learning/an-introduction-to-gradient-boosting-decision-trees/]
- 62. Comparing 13 Algorithms on 165 Datasets (hint: use ... [https://www.machinelearningmastery.com/start-with-gradient-boosting/]
- 63. DBTL Approach in Synthetic Biology [https://www.moleculardevices.com/applications/synthetic-biology/dbtl-approach]
- 64. Merging automation and fundamental discovery into the ... [https://www.sciencedirect.com/science/article/pii/S0167779922000725]
- 65. A machine learning Automated Recommendation Tool for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7519645/]
- 66. A generalized platform for artificial intelligence-powered ... [https://www.nature.com/articles/s41467-025-61209-y]
- 67. Systems biology approaches integrated with artificial ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7546651/]
- 68. Synthetic biology and metabolic engineering paving the way ... [https://pubs.rsc.org/en/content/articlehtml/2025/ya/d5ya00118h]