Stoichiometric Inconsistency and Network Gaps: From Detection to Resolution in Metabolic Models
This article explores the critical challenge of stoichiometric inconsistencies in genome-scale metabolic models (GEMs) and their role in creating network gaps that impair predictive accuracy. Aimed at researchers, scientists, and drug development professionals, it details how these structural errors arise, their impact on flux balance analysis, and the computational methods—from established algorithms like fastGapFill to emerging deep learning tools like CHESHIRE—used to detect and resolve them. The content further covers troubleshooting techniques for error isolation, the validation of gap-filling solutions, and the implications of robust, stoichiometrically consistent models for advancing biomedical research and therapeutic discovery.
Stoichiometric Inconsistency and Network Gaps: From Detection to Resolution in Metabolic Models
Abstract
This article explores the critical challenge of stoichiometric inconsistencies in genome-scale metabolic models (GEMs) and their role in creating network gaps that impair predictive accuracy. Aimed at researchers, scientists, and drug development professionals, it details how these structural errors arise, their impact on flux balance analysis, and the computational methods—from established algorithms like fastGapFill to emerging deep learning tools like CHESHIRE—used to detect and resolve them. The content further covers troubleshooting techniques for error isolation, the validation of gap-filling solutions, and the implications of robust, stoichiometrically consistent models for advancing biomedical research and therapeutic discovery.
The Root of the Problem: Defining Stoichiometric Inconsistency and Network Gaps
Stoichiometric modeling is a constraint-based methodology used to analyze metabolic networks at the genome scale, relying fundamentally on mass balance principles to predict cellular behavior without requiring detailed kinetic parameters [1]. This approach has become indispensable in systems biology for studying the systemic properties of metabolic networks, providing insight into metabolic plasticity, robustness, and an organism's ability to cope with different environments [2]. The accuracy of these models depends critically on correct stoichiometric specifications, as errors can create network gaps and structural inconsistencies that compromise predictive capability and biological relevance [3] [4].
Stoichiometric models bridge the gap between genomic information and metabolic functionality, enabling researchers to predict metabolic flux distributions, identify essential genes, and pinpoint thermodynamic constraints [1]. In pharmaceutical and biomedical research, these models are particularly valuable for drug target identification, understanding disease mechanisms, and optimizing bioproduction processes [4]. The fundamental principle governing all stoichiometric modeling is mass conservation, which requires that atoms are neither created nor destroyed in biochemical reactions [2] [3].
Mathematical Foundations of Stoichiometric Modeling
The Stoichiometric Matrix and Mass Balance
The cornerstone of stoichiometric modeling is the stoichiometric matrix (denoted as N), which mathematically represents the metabolic network structure [2] [1]. This m × n matrix contains the stoichiometric coefficients of m metabolites participating in n reactions, where each element nij represents the stoichiometric coefficient of metabolite i in reaction j [2].
The rate of change of metabolite concentrations is described by the system of ordinary differential equations:
dx/dt = N · v
where x is the m-dimensional metabolite concentration vector and v is the n-dimensional reaction rate vector [2]. At steady state (a fundamental assumption in most stoichiometric analyses), the time derivatives become zero, reducing the equation to:
N · v = 0
This equation represents the mass balance constraint for each metabolite in the network, indicating that the total production and consumption rates for each metabolite must be equal [2] [1].
Chemical Moisty Conservation
In addition to mass balance, metabolic networks exhibit chemical moiety conservation, where certain chemical groups (e.g., adenosine, phosphate) are conserved within the network [2]. These conservation relationships impose additional constraints on the system and can be expressed mathematically as:
L₀ · x = t
where Lâ‚€ is the moiety conservation matrix, x is the metabolite concentration vector, and t is the vector of total moiety concentrations [2]. These relationships allow for the decomposition of metabolites into dependent and independent sets, reducing the system's complexity.
Table 1: Key Mathematical Components in Stoichiometric Modeling
| Component | Symbol | Description | Role in Modeling |
|---|---|---|---|
| Stoichiometric Matrix | N | m × n matrix of stoichiometric coefficients | Defines network structure and mass balance constraints |
| Flux Vector | v | n-dimensional vector of reaction rates | Represents metabolic activity state |
| Metabolite Vector | x | m-dimensional vector of metabolite concentrations | Defines metabolic pool sizes |
| Kernel Matrix | K | Null-space matrix of N | Contains all steady-state flux solutions |
| Moiety Matrix | Lâ‚€ | Conservation relationship matrix | Defines conserved chemical groups |
Methodologies and Analytical Approaches
Core Stoichiometric Modeling Techniques
Several computational methodologies have been developed within the stoichiometric modeling framework, each with distinct purposes and mathematical implementations [1].
Flux Balance Analysis (FBA) is a widely used constraint-based approach that predicts metabolic flux distributions by optimizing an objective function (e.g., biomass production, ATP synthesis) subject to stoichiometric constraints [2] [5]. FBA formulates metabolism as a linear programming problem:
Maximize cᵀ · v subject to N · v = 0 and α ≤ v ≤ β
where c is the vector of objective coefficients, and α and β are lower and upper bounds on fluxes [1] [5].
Metabolic Flux Analysis (MFA) utilizes measured extracellular fluxes in combination with the stoichiometric model to determine intracellular fluxes that cannot be directly measured [4] [1]. The flux estimation is typically performed as a weighted least-squares problem:
Minimize ‖(rout - rin) - S · v‖² subject to α ≤ v ≤ β
where rout and rin are measured external metabolite excretion and uptake rates [4].
Network-Based Pathway Analysis identifies systemic properties of metabolic networks by analyzing the set of pathways through the network [1]. This includes methods like Elementary Flux Modes (EFMs) and Extreme Pathways (ExPas), which represent minimal sets of reactions that can operate at steady state [2].
Protocols for Metabolic Network Analysis
Protocol 1: Stoichiometric Model Construction and Validation
- Network Reconstruction: Compile all metabolic reactions from genome annotation databases and biochemical literature [4]
- Stoichiometric Matrix Assembly: Construct the N matrix with metabolites as rows and reactions as columns [2]
- Mass Balance Verification: Check that each reaction is atomically balanced using atomic mass analysis (AMA) [3]
- Moiety Conservation Analysis: Identify conserved chemical moieties using left null-space analysis of N [2]
- Gap Filling: Identify and fill network gaps using biochemical databases and computational tools like OptFill [6]
- Model Validation: Compare predictions with experimental data (e.g., gene essentiality, growth rates) [4]
Protocol 2: Flux Balance Analysis Implementation
- Objective Function Definition: Select appropriate biological objective (e.g., biomass maximization) [5]
- Constraint Specification: Define environmental conditions through exchange reaction bounds [1]
- Linear Programming Solution: Solve the optimization problem using algorithms like simplex or interior point methods [5]
- Solution Space Characterization: Use tools like CoPE-FBA to comprehensively enumerate optimal flux spaces [5]
- Flux Variability Analysis: Determine the range of possible fluxes for each reaction while maintaining optimal objective value [2] [5]
- Validation with Experimental Data: Compare predictions with measured flux data, if available [4]
Table 2: Common Stoichiometric Modeling Methods and Applications
| Method | Mathematical Basis | Primary Application | Key Output |
|---|---|---|---|
| Flux Balance Analysis (FBA) | Linear Programming | Prediction of optimal flux distributions | Optimal flux vector and objective value |
| Metabolic Flux Analysis (MFA) | Least-Squares Regression | Determination of intracellular fluxes from extracellular measurements | Complete flux map with confidence intervals |
| Flux Variability Analysis (FVA) | Linear Programming | Determination of flux ranges in optimal states | Minimum and maximum flux for each reaction |
| Elementary Flux Modes (EFM) | Convex Analysis | Identification of minimal functional pathways | Set of irreducible steady-state flux distributions |
| Comprehensive Polyhedra Enumeration (CoPE-FBA) | Polyhedral Geometry | Complete characterization of optimal flux spaces | Vertices, rays, and linealities of flux polyhedron |
Stoichiometric Inconsistency and Network Gaps
Stoichiometric inconsistencies represent a critical class of errors in metabolic models that can create network gaps and compromise predictive accuracy [3]. These inconsistencies arise when the stoichiometric constraints imply that one or more chemical species must have zero mass, indicating fundamental problems in network structure [3].
The primary types of stoichiometric inconsistencies include:
- Mass Balance Errors: Discrepancies between total mass of reactants and products in individual reactions [3]
- Moiety Balance Errors: Imbalances in chemical structures (e.g., phosphate groups) between reactants and products [3]
- Stoichiometric Inconsistencies: Structural errors where reaction network topology implies impossible mass relationships [3]
- Thermodynamically Infeasible Cycles (TICs): Cyclic reaction sets that could theoretically operate without energy input [6]
Detection and Resolution Methods
Algorithm 1: Moiety Balance Analysis Moiety analysis detects imbalances of chemical moieties using the same mathematical framework as atomic mass analysis but operates in units of moieties rather than individual atoms [3]. This approach is particularly valuable for detecting errors involving chemical groups with slightly different atomic formulas in different molecular contexts.
Algorithm 2: Graphical Analysis of Mass Equivalence Sets (GAMES) GAMES isolates stoichiometric inconsistencies by identifying small subsets of reactions and species (Reaction Isolation Set - RIS and Species Isolation Set - SIS) that explain structural errors [3]. This method simplifies error remediation by pinpointing the specific network elements requiring correction.
Structural Error Isolation Workflow
Advanced Topics and Research Directions
Gapfilling and Model Correction
Gapfilling is the process of identifying and resolving network gaps in metabolic reconstructions [6]. Current approaches use databases of biochemical functionalities to address gaps on a per-metabolite basis but often struggle with creating thermodynamically infeasible cycles (TICs) [6]. Advanced methods like OptFill perform holistic, TIC-avoiding whole-model gapfilling through optimization-based multi-step procedures [6].
The OptFill methodology involves:
- Identifying network gaps through connectivity analysis
- Proposing candidate reactions from biochemical databases
- Selecting minimal reaction sets that restore functionality
- Ensuring thermodynamic feasibility by avoiding TICs
- Validating added reactions against experimental data [6]
Standardization Challenges in Metabolic Models
A significant challenge in stoichiometric modeling, particularly for human metabolic networks, is the lack of standardization in reconstruction methods, representation formats, and model repositories [4]. This hinders direct comparison between models, selection of appropriate models for specific applications, and understanding of how metabolic network reconstructions evolve [4].
Standardization efforts focus on:
- Consistent annotation of genes, proteins, and reactions
- Uniform representation of compartmentalization
- Standardized formats for model exchange
- Benchmarks for model quality assessment [4]
Stoichiometric Modeling Pipeline
Table 3: Key Resources for Stoichiometric Modeling Research
| Resource | Type | Primary Function | Application Context |
|---|---|---|---|
| COBRA Toolbox | Software Package | Constraint-based reconstruction and analysis | MATLAB-based suite for FBA, MFA, and model validation [3] |
| MEMOTE | Software Tool | Model testing and validation | Automated quality assessment of genome-scale models [3] |
| OptFill | Gapfilling Algorithm | TIC-avoiding model completion | Holistic gapfilling of stoichiometric models [6] |
| BioModels | Model Repository | Curated model database | Source of validated biochemical models [3] |
| SBMLLint | Linting Tool | Structural error detection | Identification of mass balance and moiety errors [3] |
| CoPE-FBA | Analysis Method | Comprehensive flux space enumeration | Complete characterization of optimal FBA solutions [5] |
Stoichiometric modeling provides a powerful framework for analyzing metabolic networks based on fundamental mass balance principles. The accuracy of these models depends critically on avoiding stoichiometric inconsistencies, which can create network gaps and compromise predictive capability. Advanced methods for error detection, gapfilling, and solution space characterization continue to enhance the biological relevance and predictive power of stoichiometric models.
As the field advances, standardization of reconstruction methods, representation formats, and model repositories will be essential for enabling direct comparison between models and consistent integration of multi-omic data. These developments will further solidify the role of stoichiometric modeling as an indispensable tool in systems biology, metabolic engineering, and pharmaceutical research.
Stoichiometric inconsistency represents a fundamental error in the specification of biochemical reaction networks, violating the universal constraint that mass is conserved in every chemical transformation [3] [7]. In systems biology, particularly in stoichiometric modeling of metabolism, these inconsistencies arise when the total mass of atoms in the reactants does not equal the total mass of atoms in the products of a reaction [3]. This error violates the principle of conservation of mass, where molecular masses are always positive, and on each side of a reaction, mass must be conserved [7].
A single incorrectly defined reaction can lead to stoichiometric inconsistency throughout an entire model, resulting in unconserved metabolites [7]. These inconsistencies create profound problems in computational models, as they may give rise to thermodynamically infeasible cycles that either produce mass from nothing or consume mass from the model [7]. The presence of such errors undermines the predictive accuracy of metabolic models and can lead to biologically impossible predictions, such as the existence of metabolites with effectively zero mass [3].
The growing complexity of reaction-based models in systems biology necessitates early detection and resolution of these fundamental errors [3]. As biochemical networks in repositories like BioModels now range from tens to thousands of reactions, with over 800 curated models available, the correctness of these models is of particular concern since they often serve as starting points for new research [3]. Understanding and addressing stoichiometric inconsistencies is therefore essential for reliable metabolic modeling in biomedical research and drug development.
Fundamental Concepts and Biochemical Principles
Mass Balance vs. Moiety Balance
In biochemical modeling, two complementary concepts of balance must be considered:
Atomic Mass Balance: This fundamental approach compares the counts of individual atoms in reactants and products [3]. Implemented through Atomic Mass Analysis (AMA), it requires annotations of chemical species to obtain atomic formulas and looks for differences in atoms between reactants and products [3]. This method can check both charge balance and mass balance when atom ionization states are specified [3].
Moiety Balance: A moiety represents "a part or portion of a molecule, generally complex, having a characteristic chemical or pharmacological property" [3]. Unlike individual atoms, a single moiety may refer to groupings of atoms that have slightly different atomic formulas, such as the inorganic phosphate moiety found in ATP, ADP, and free Pi [3]. Moiety-preserving reactions are exceedingly common in biochemistry, particularly in transferase reactions that facilitate the transfer of chemical groups between molecules [3].
The critical distinction emerges when considering reactions like ATP hydrolysis, commonly written as ATP → ADP + Pi [3]. While this reaction is moiety balanced (one adenosine and three phosphates on both sides), it is not mass balanced due to differences in the atomic formulas of the inorganic phosphates in different molecular contexts [3]. To achieve mass balance, water must be included as a reactant, yet many modelers omit such "implicit molecules" whose concentrations remain relatively constant in solution [3].
Types of Structural Errors in Reaction Networks
Stoichiometric inconsistencies manifest through several specific structural errors in biochemical networks:
Mass Balance Errors: Discrepancies between the total mass of reactants and products [3]. These are detectable through AMA when complete atomic formulas are available [3].
Stoichiometric Inconsistency: A structural error implying that one or more chemical species have a mass of zero [3]. This type of error can propagate through networks, creating logical contradictions where a metabolite's mass must simultaneously be larger than itself [3].
Moiety Balance Errors: Imbalances of chemical structures between reactants and products that cannot be detected through atomic-level analysis alone [3]. These occur when reactions that should preserve moiety counts are incorrectly specified.
Table 1: Comparison of Balance Types in Biochemical Networks
| Balance Type | Analysis Method | Key Principle | Common Examples |
|---|---|---|---|
| Atomic Mass Balance | Atomic Mass Analysis (AMA) | Conservation of individual atom counts | Complete combustion reactions; oxidation processes |
| Charge Balance | Atomic Mass Analysis with ionization states | Conservation of electrical charge | Ion transport; electron transfer chains |
| Moiety Balance | Moiety Analysis | Conservation of functional chemical groups | Phosphate transfer (kinases); methyl group transfer |
Detection Methodologies and Algorithms
Computational Frameworks for Consistency Checking
Multiple algorithmic approaches have been developed to detect stoichiometric inconsistencies in biochemical networks:
Stoichiometric Consistency Analysis: This test uses an implementation of the algorithm presented by Gevorgyan et al. (2008) to detect stoichiometric inconsistencies [7] [8]. The method identifies unconserved metabolites using the algorithm described in section 3.2 of the same publication [7]. In practical applications, this approach can reveal significant issues, with some models containing over 60% unconserved metabolites [8].
Moiety Analysis Algorithm: This approach adapts the same algorithmic framework as AMA but operates in units of moieties rather than atomic masses [3]. This enables detection of chemical structure imbalances that would be missed by atomic-level analysis alone [3].
Linear Programming Analysis: This method detects stoichiometric inconsistencies through optimization approaches that identify violations of mass conservation constraints [3].
The memote consistency test suite provides a comprehensive implementation of these methodologies, testing for stoichiometric consistency, unconserved metabolites, inconsistent minimal stoichiometries, and energy-generating cycles [7].
Error Isolation Techniques
Advanced methods have been developed not only to detect inconsistencies but to isolate their sources:
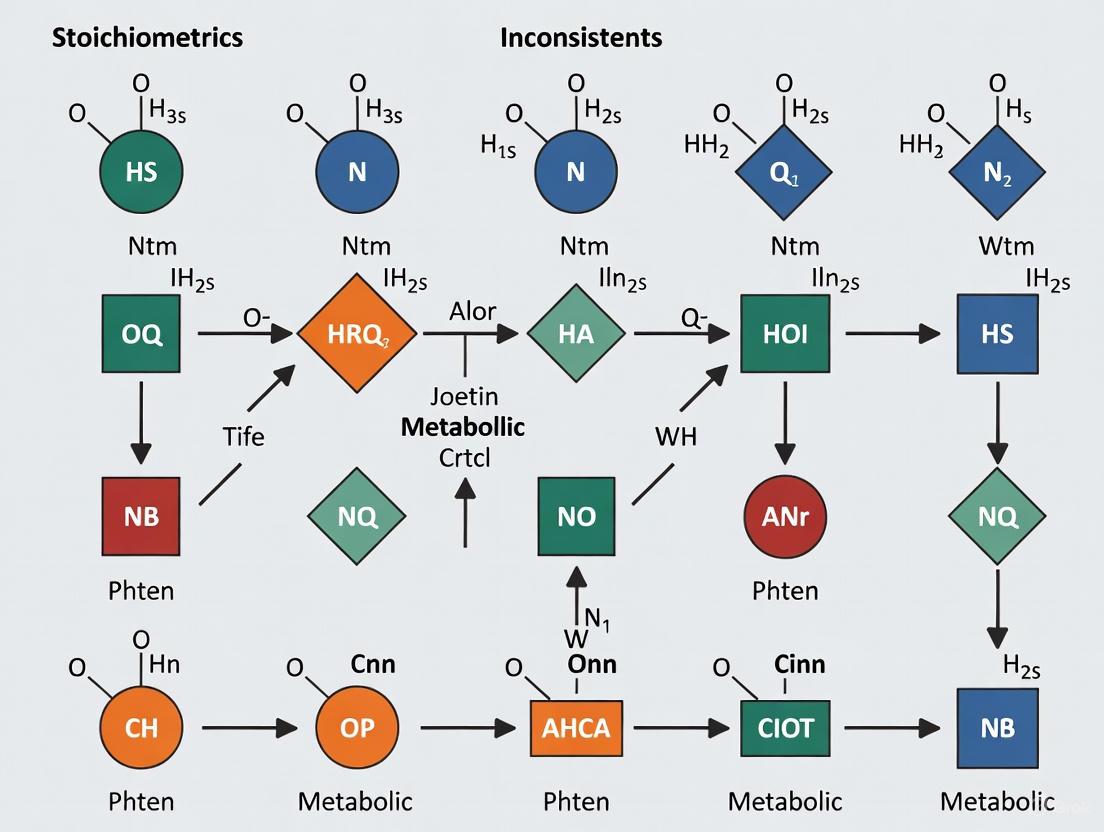
Graphical Analysis of Mass Equivalence Sets (GAMES): This algorithm provides isolation for stoichiometric inconsistencies by constructing explanations that relate errors in network structure to specific elements of the reaction network [3]. It identifies Reaction Isolation Sets (RIS) and Species Isolation Sets (SIS) that pinpoint the reactions and species causing errors [3].
Comprehensive Polyhedra Enumeration Flux Balance Analysis (CoPE-FBA): This approach characterizes the complete optimal flux space of stoichiometric models, revealing how a few subnetworks shape the geometry of optimal FBA solutions [5]. The method shows that typically only 5-10% of all reactions in a network determine the solution space [5].
The error isolation process involves identifying computationally simple explanations that show how the RIS and SIS cause the error, enabling researchers to efficiently remediate model errors [3].
Diagram 1: Stoichiometric Consistency Checking Workflow. This diagram illustrates the sequential process for detecting and isolating stoichiometric inconsistencies in biochemical models, incorporating multiple analysis methods and error isolation techniques.
Impact on Metabolic Network Analysis
Consequences for Metabolic Modeling
Stoichiometric inconsistencies create profound challenges for metabolic network analysis and prediction:
Thermodynamically Infeasible Cycles: Inconsistent models may give rise to cycles that either produce mass from nothing or consume mass from the model [7]. These include Energy Generating Cycles that provide reduced metabolites without requiring nutrient uptake, potentially increasing predicted growth rates by up to 25% in FBA, making growth predictions unreliable [7].
Blocked Reactions and Network Gaps: Universally blocked reactions cannot carry any flux when all model boundaries are open, typically caused by network gaps attributed to scope or knowledge limitations [7]. Orphan metabolites (only consumed) and dead-end metabolites (only produced) indicate structural network problems and knowledge gaps [7].
Flux Balance Analysis Limitations: Inconsistent models compromise FBA predictions, as the solution space becomes distorted by stoichiometric errors [5]. The presence of even a few inconsistent reactions can dramatically expand the feasible solution space with biologically impossible flux distributions.
Network-Wide Implications
The impact of stoichiometric inconsistencies extends throughout metabolic networks:
Solution Space Distortion: CoPE-FBA analysis demonstrates that optimal flux spaces of genome-scale stoichiometric models are determined by a few subnetworks [5]. When these subnetworks contain stoichiometric inconsistencies, the entire solution space becomes compromised.
Flux-Concentration Duality Breakdown: Under normal conditions, mathematical modeling of biochemical networks can be equivalently described in terms of either concentrations or unidirectional fluxes [9]. Stoichiometric inconsistencies disrupt this duality, preventing equivalent descriptions using these different perspectives.
Multi-omic Integration Challenges: Inconsistent metabolic models hinder integration with other biological data layers, such as transcriptomic and proteomic data, limiting their utility in systems biology approaches [4] [10].
Table 2: Common Structural Errors in Biochemical Networks and Their Impacts
| Error Type | Detection Method | Impact on Model Predictions | Remediation Approaches |
|---|---|---|---|
| Unconserved Metabolites | Stoichiometric consistency test [7] | Mass can be created/destroyed; thermodynamic infeasibility | Add missing reactants/products; verify formulas |
| Energy Generating Cycles | Detect energy metabolite production from nothing [7] | Artificial ATP production; inflated growth predictions | Add thermodynamic constraints; verify reaction directions |
| Blocked Reactions | Flux Variability Analysis with open exchanges [7] | Limited network functionality; incomplete pathway coverage | Gap-filling algorithms; add missing transport reactions |
| Orphan Metabolites | Structural analysis of reaction equations [7] | Metabolites only consumed; accumulation impossible | Add producing reactions; verify compartmentalization |
| Dead-end Metabolites | Structural analysis of reaction equations [7] | Metabolites only produced; depletion impossible | Add consuming reactions; verify degradation pathways |
Research Reagent Solutions and Experimental Tools
Table 3: Essential Research Tools for Stoichiometric Consistency Analysis
| Tool/Resource | Primary Function | Application Context | Key Features |
|---|---|---|---|
| SBMLLint [3] | Open-source linting for SBML models | Structural error detection in reaction networks | Moiety analysis; GAMES for error isolation; MIT license |
| MEMOTE [7] [8] | Test suite for stoichiometric consistency | Comprehensive model quality assessment | Implements Gevorgyan et al. algorithm; consistency scoring |
| COBRA Toolbox [3] | Constraint-based reconstruction and analysis | Genome-scale metabolic modeling | Atomic mass analysis with R-groups; charge balance checking |
| OptFill [6] | Optimization-based gapfilling | Holistic, infeasible cycle-free model completion | Avoids thermodynamically infeasible cycles during gapfilling |
| CoPE-FBA [5] | Comprehensive polyhedra enumeration | Complete characterization of optimal flux spaces | Identifies subnetworks determining solution space geometry |
Advanced Research Applications and Case Studies
Real-World Implications in Metabolic Research
The practical significance of stoichiometric consistency is evident across multiple research domains:
Stoichiometric Balance in Protein Networks: Research integrating protein copy numbers with interaction networks has established a Stoichiometric Balance Ratio (SBR) to quantify whether each protein in a network has abundance that is sub- or super-stoichiometric relative to global competition for binding [11]. This approach reveals how highly abundant proteins like clathrin are super-stoichiometric, while variations in both abundance and unique binding networks create widespread competition for shared binding sites [11].
Gene Expression Integration Challenges: Studies integrating gene expression profiles with metabolic pathways reveal substantial inconsistencies between expression data and anticipated network dynamics [10]. The Inconsistency Index (I) quantifies disagreement between expression data and network objectives, while Metabolic Coherence (MC) measures coordinated expression of connected reaction structures [10]. These measures show strong anticorrelation, demonstrating that inconsistencies between metabolic processes and gene expression can be understood from a network perspective [10].
Polymer Science Applications: Beyond metabolic networks, stoichiometric principles critically influence material properties in polymer science, where controlling functional group stoichiometry and crosslinking density determines reprocessability in covalent adaptable networks [12]. Precise stoichiometric design enables tuning of viscoelastic properties and mechanical behavior in polymer systems [12].
Protocol for Consistency Testing
The memote test suite provides a standardized protocol for stoichiometric consistency assessment:
Stoichiometric Consistency Test: Apply the algorithm from Gevorgyan et al. to verify overall model consistency [7].
Unconserved Metabolite Identification: Use the section 3.2 algorithm from the same paper to identify all unconserved metabolites [7].
Energy Generating Cycle Detection: Implement the Fritzemeier et al. algorithm to identify cycles that produce energy metabolites from nothing [7].
Charge and Mass Balance Verification: Check all non-boundary reactions for charge and mass balance, excluding reactions with missing formula or charge annotations [7].
Structural Network Analysis: Identify orphan metabolites, dead-ends, and disconnected metabolites through structural analysis of reaction equations [7].
Diagram 2: Model Consistency Assessment Protocol. This workflow outlines the standardized procedure for evaluating stoichiometric consistency in biochemical models, from data extraction through comprehensive testing and reporting.
Stoichiometric inconsistencies represent a critical challenge in biochemical network modeling, creating mass imbalance errors that propagate through computational models and compromise their predictive accuracy. These inconsistencies manifest as unconserved metabolites, energy-generating cycles, and stoichiometric contradictions that violate fundamental physical principles [3] [7].
Within the broader context of network gap research, stoichiometric inconsistencies create profound limitations by introducing structural errors that distort the feasible solution space of metabolic models [5]. These errors hinder the integration of multi-omic data layers [4] [10], compromise flux balance predictions [5], and create thermodynamic impossibilities that render models biologically implausible [7].
Advanced detection methodologies, including moiety analysis [3], GAMES for error isolation [3], and comprehensive consistency testing frameworks [7], provide researchers with powerful tools to identify and remediate these issues. The development of stoichiometric balance metrics across biological scales [11] and the application of stoichiometric principles in diverse fields [12] underscore the fundamental importance of mass conservation in predictive biological modeling.
As biochemical networks continue to increase in complexity and scope, maintaining stoichiometric consistency remains essential for developing accurate, predictive models that can reliably inform drug development, metabolic engineering, and biomedical research. The integration of robust consistency checking throughout the model development lifecycle represents a critical step toward realizing the full potential of systems biology in therapeutic applications.
Genome-scale Metabolic models (GEMs) are powerful computational tools that provide a mathematical representation of an organism's metabolism, mapping the complex network of biochemical reactions [13]. They are indispensable in advancing disciplines such as metabolic engineering, microbial ecology, and drug discovery. However, the presence of knowledge gaps—missing reactions due to incomplete genomic and functional annotations—represents a significant challenge to model accuracy and utility. These gaps often manifest as stoichiometric inconsistencies, disrupting the flow of metabolites through the network and creating "dead-end" metabolites that cannot be produced or consumed [13]. This article explores how these stoichiometric inconsistencies create network gaps and, consequently, how such gaps propagate through computational analyses to produce flux errors and potentially false biological insights, with a particular focus on implications for drug development and biomedical research.
Quantifying the Gap-Filling Performance of Computational Methods
The performance of different computational methods in addressing network gaps can be systematically evaluated. The following table summarizes the core abilities of various topology-based gap-filling methods, highlighting their distinct approaches and limitations.
Table 1: Comparison of Topology-Based Gap-Filling Methods for Metabolic Models
| Method Name | Core Methodology | Key Advantages | Documented Limitations |
|---|---|---|---|
| CHESHIRE (2023) [13] | Deep learning using Chebyshev spectral graph convolutional networks on metabolic hypergraphs. | Superior prediction accuracy; does not require phenotypic data for training; scalable to large reaction pools. | Performance may vary with network size and completeness. |
| Neural Hyperlink Predictor (NHP) [13] | Neural network that approximates hypergraphs using graphs for node feature generation. | Separates candidate reactions from training. | Loss of higher-order information due to graph approximation; less accurate than CHESHIRE. |
| C3MM [13] | Clique Closure-based Coordinated Matrix Minimization. | Integrated training-prediction process. | Limited scalability; model must be re-trained for each new reaction pool. |
| Marginal Distribution Sampling (MDS) [14] | Fills gaps using mean available values measured under similar meteorological conditions (primarily for EC data). | Standardized method used in FLUXNET and ICOS. | Systematically overestimates CO₂ emissions at northern sites (>60° latitude) due to skewed radiation distributions. |
Quantitative validation is critical for establishing the reliability of these methods. In an internal validation test designed to evaluate the ability to recover artificially removed reactions, CHESHIRE demonstrated superior performance. The test involved 108 BiGG models and 818 AGORA models, with reactions split into training and testing sets over 10 Monte Carlo runs [13].
Table 2: Internal Validation Performance on Artificial Gaps
| Performance Metric | CHESHIRE | NHP | C3MM | Node2Vec-Mean (NVM) |
|---|---|---|---|---|
| Area Under the Curve (AUC) | Outperformed other methods [13] | Lower than CHESHIRE | Lower than CHESHIRE | Used as a baseline; lower than other methods |
| Key Differentiator | Exploits a sophisticated CSGCN and Frobenius norm-based pooling [13]. | Lacks higher-order information capture [13]. | Lacks scalability; requires re-training for new pools [13]. | Simple architecture without feature refinement [13]. |
Furthermore, an external validation assessed the impact of gap-filling on predicting metabolic phenotypes. Using 49 draft GEMs from CarveMe and ModelSEED pipelines, CHESHIRE improved the theoretical predictions of fermentation product and amino acid secretion [13]. This demonstrates that advanced gap-filling can directly enhance the functional utility of metabolic models.
Experimental Protocols for Method Validation
Internal Validation Protocol: Recovering Artificially Introduced Gaps
This protocol tests a method's ability to reconstruct a known, complete network by intentionally creating and then filling gaps [13].
- Input Preparation: Obtain a high-quality, curated GEM.
- Data Splitting: Split the metabolic reactions of the GEM into a training set (e.g., 80%) and a testing set (e.g., 20%). Perform this split over multiple (e.g., 10) Monte Carlo runs to ensure statistical robustness [13].
- Negative Sampling: Generate negative (non-existent) reactions for both training and testing sets at a 1:1 ratio to positive reactions. This is typically done by replacing half of the metabolites in a positive reaction with randomly selected metabolites from a universal metabolite pool [13].
- Model Training: Train the gap-filling method (e.g., CHESHIRE, NHP) using the combined set of positive and negative reactions in the training set.
- Performance Testing: Apply the trained model to the testing set mixed with its derived negative reactions. The model predicts a confidence score for each reaction in this test pool.
- Evaluation: Calculate performance metrics, such as Area Under the Curve (AUC), by comparing the model's predictions against the ground truth (i.e., which reactions were originally removed and which negative reactions are fake) [13].
External Validation Protocol: Predicting Metabolic Phenotypes
This protocol validates the method's real-world utility by testing its impact on the model's predictive functionality [13].
- Model Selection: Select a set of draft GEMs that have been reconstructed from genomic data using standard pipelines (e.g., CarveMe, ModelSEED).
- Gap-Filling: Apply the gap-filling method to these draft models, adding a set of candidate reactions predicted to be missing.
- Phenotypic Prediction: Use the original and the gap-filled models to simulate specific metabolic phenotypes (e.g., secretion of fermentation products, amino acid auxotrophy).
- Validation against Data: Compare the simulation results against known experimental data (e.g., from culture studies) for the organism.
- Evaluation: Quantify the improvement in prediction accuracy (e.g., increase in true positives, reduction in false negatives) in the gap-filled models compared to the original draft models [13].
Protocol for Assessing Carbon Balance Errors in Eddy Covariance Data
While not specific to GEMs, this protocol from flux measurement science exemplifies a robust validation workflow relevant to gap-filling in time-series data [14].
- Synthetic Data Generation: Create a synthetic, full time series of COâ‚‚ fluxes that corresponds to the observed fluxes at a site, ensuring a known "true" carbon balance [14].
- Introduction of Artificial Gaps: Introduce realistic artificial gaps (in both length and timing) into the synthetic data set. Common gap levels are 30%, 50%, and 70% of data [14].
- Gap-Filling: Apply the gap-filling methods (e.g., MDS, XGBoost) to the gapped synthetic data.
- Error Calculation: Calculate the annual carbon balance from the gap-filled time series. The balance error is determined as the difference between this estimated balance and the known true balance of the synthetic data [14].
Diagram 1: Internal Validation Workflow
Table 3: Essential Computational Tools and Databases for Metabolic Model Gap-Filling
| Resource Name | Type | Primary Function in Gap-Filling |
|---|---|---|
| BiGG Models [13] | Knowledgebase | A repository of high-quality, curated GEMs; used as a gold-standard benchmark for testing and validating gap-filling methods. |
| AGORA Models [13] | Knowledgebase | A resource of genome-scale metabolic reconstructions for human gut microbes; provides a diverse set of models for validation. |
| CarveMe [13] | Software Tool | An automated pipeline for draft model reconstruction from genomic data; produces draft models that often require subsequent gap-filling. |
| ModelSEED [13] | Software Tool | A framework for the automated reconstruction and analysis of metabolic models; generates draft models that can be used for gap-filling validation. |
| REddyProc [14] | Software Tool | A tool for gap-filling eddy covariance data, implementing the MDS method; highlights domain-specific challenges in gap-filling. |
| Universal Metabolite Pool [13] | Data Resource | A comprehensive collection of known metabolites; used to generate plausible negative reactions during machine learning model training. |
| XGBoost [14] | Software Library | A machine learning library implementing gradient boosting; used as an advanced alternative to MDS for flux data gap-filling. |
Consequences of Inadequate Gap-Filling: From Bias to False Insights
Inadequate gap-filling methods can introduce systematic biases that compromise the validity of model predictions. A critical example comes from the field of eddy covariance, where the widely used Marginal Distribution Sampling (MDS) method has been shown to create significant carbon balance errors for northern sites (latitude >60°) [14]. The underlying cause is a skewed radiation distribution at high latitudes. During gap-filling, MDS samples more data from the lower range of the radiation distribution, which corresponds to underestimated photosynthetic uptake. This leads to a systematic overestimation of COâ‚‚ emissions from carbon sources and an underestimation of COâ‚‚ sequestration by carbon sinks [14]. The median balance error with MDS can range from 2–10 g C mâ»Â² yâ»Â¹ at a 30% gap level to 3–17 g C mâ»Â² yâ»Â¹ at a 70% gap level, with some errors exceeding 30 g C mâ»Â² yâ»Â¹ [14]. This demonstrates how a widely trusted method can produce predictable, directional errors under specific conditions.
In metabolic model analysis, gaps caused by stoichiometric inconsistencies prevent models from simulating known metabolic functions, leading to false-negative predictions. For instance, a draft model might incorrectly predict that an organism cannot synthesize an essential amino acid or produce a key fermentation product due to a missing reaction in an otherwise complete pathway [13]. Conversely, an inappropriate gap-filling technique might introduce reactions that create thermodynamically infeasible loops or bypass key regulatory steps, allowing the model to produce a metabolite without the necessary biochemical constraints and potentially leading to false-positive predictions. These inaccuracies can directly impact drug discovery efforts. For example, targeting an enzyme that is part of a pathway predicted to be essential in a pathogen—when in reality the pathway is non-functional or can be bypassed due to model gaps—could lead to failed therapeutic strategies. Thus, robust gap-filling is not merely a technical exercise but a critical step in ensuring the biological relevance and predictive power of in-silico models.
Diagram 2: Impact of Network Gaps on Predictions
The impact of gaps on model predictions is profound and far-reaching, leading to everything from quantifiable flux errors to fundamentally flawed biological insights. Stoichiometric inconsistencies create network gaps that disrupt the biochemical logic of metabolic models, while inadequate gap-filling methods can introduce systematic biases, as evidenced by the performance of MDS in environmental flux data and the limitations of early machine learning methods for GEMs. The development of advanced, topology-based methods like CHESHIRE, which leverage deep learning on hypergraph representations of metabolism, offers a promising path forward. By providing more accurate and scalable gap-filling, these tools can significantly improve the predictive fidelity of models. For researchers in drug development and biomedical science, relying on models refined by such robust methods is becoming increasingly critical to generate reliable hypotheses, identify valid therapeutic targets, and avoid the costly dead ends that stem from false biological insights.
Stoichiometric inconsistency in chemical and biological networks creates critical knowledge gaps that hinder the prediction of synthesizable materials and the understanding of metabolic processes. This whitepaper explores how imbalances in elemental composition disrupt network connectivity and functionality. We examine advanced computational methods, including machine learning and hypergraph-based approaches, that identify and rectify these stoichiometric gaps. By integrating data from materials science and metabolic network analysis, this guide provides researchers with robust protocols for predicting synthesizability and filling network gaps, ultimately accelerating discovery in drug development and materials design.
In both inorganic materials science and cellular biochemistry, the balanced representation of elemental composition—stoichiometry—is fundamental for predicting stable compounds and viable metabolic pathways. Stoichiometric inconsistency refers to imbalances in elemental representation that lead to network gaps, disrupting the connectivity and functionality of chemical reaction networks (CRNs) and genome-scale metabolic models (GEMs). These gaps manifest as dead-end metabolites that cannot be produced or consumed, or as computationally predicted compounds that are experimentally unsynthesizable [15] [16].
The challenge extends beyond simple atomic mass balance to the identification of chemically plausible linkages between moieties. In metabolic engineering, incomplete genomic and functional annotations result in GEMs with missing reactions, creating unrealistic metabolic predictions [16]. Similarly, in materials science, the majority of candidate materials identified through high-performance computing are impractical to synthesize due to intricate synthesis constraints [15]. Understanding and resolving these stoichiometric inconsistencies is therefore critical for advancing predictive capabilities in both fields.
Computational Frameworks for Gap Analysis and Prediction
Machine Learning for Synthesizability Prediction
Positive-unlabeled learning represents a powerful machine learning approach for predicting the synthesizability of inorganic material stoichiometries. This method addresses the challenge where only positive (synthesizable) examples are definitively known, while unsynthesizable compounds remain unlabeled.
Experimental Protocol for Synthesizability Prediction:
- Data Collection: Compile a database of known synthesizable inorganic compositions from crystallographic databases.
- Feature Initialization: Encode each elemental stoichiometry using compositional descriptors and structural features.
- Model Training: Apply positive-unlabeled learning algorithms where known synthesizable compositions serve as positive examples, while random stoichiometries from chemical space serve as unlabeled examples.
- Validation: Assess model performance using recall and precision metrics against held-out test sets of known materials.
- Experimental Guidance: Use model predictions with high confidence scores to guide exploration of new compositional spaces, such as the discovery of the new quaternary oxide phase Cuâ‚„FeV₃Oâ‚₃ [15].
This approach has demonstrated a true positive rate of 83.4% and an estimated precision of 83.6% on test datasets, enabling the construction of continuous synthesizability phase maps that agree with available synthetic data [15].
Hypergraph Learning for Metabolic Network Gap-Filling
Metabolic networks naturally form hypergraphs where reactions (hyperedges) connect multiple metabolite nodes simultaneously. The CHESHIRE (CHEbyshev Spectral HyperlInk pREdictor) method uses deep learning on hypergraph representations of GEMs to predict missing reactions purely from network topology, without requiring experimental phenotypic data [16].
Table 1: Performance Comparison of Topology-Based Gap-Filling Methods
| Method | Architecture | AUROC (Mean) | Key Innovation |
|---|---|---|---|
| CHESHIRE | Chebyshev Spectral Graph Convolutional Network | 0.92 | Hypergraph learning with feature refinement |
| NHP | Graph-based approximation of hypergraphs | 0.85 | Neural network with mean pooling |
| C3MM | Clique Closure-based Matrix Minimization | 0.79 | Integrated training-prediction process |
| Node2Vec-mean | Random walk graph embedding | 0.74 | Simple baseline with mean pooling |
Experimental Protocol for CHESHIRE:
- Network Representation: Convert the metabolic network into a hypergraph where each reaction is a hyperlink connecting all participating metabolites.
- Feature Initialization: Generate initial feature vectors for each metabolite from the hypergraph incidence matrix using an encoder-based neural network.
- Feature Refinement: Apply Chebyshev Spectral Graph Convolutional Network (CSGCN) on a decomposed graph to refine metabolite features by incorporating information from connected metabolites.
- Pooling and Scoring: Use maximum minimum-based and Frobenius norm-based pooling functions to integrate metabolite features into reaction-level representations, then score reaction existence probability.
- Internal Validation: Test the method by artificially removing reactions from high-quality GEMs (e.g., 108 BiGG models) and measuring recovery accuracy.
- External Validation: Assess improved prediction of metabolic phenotypes (e.g., fermentation products, amino acid secretion) in 49 draft GEMs [16].
CHESHIRE's architecture enables it to outperform other topology-based methods in recovering artificially removed reactions and improves phenotypic predictions for draft metabolic models [16].
Visualization of Chemical Space and Reaction Networks
Chemical Space Networks (CSNs)
Chemical Space Networks provide powerful visualizations for exploring relationships between chemical moieties. In CSNs, compounds are represented as nodes connected by edges defined by pairwise relationships such as 2D fingerprint Tanimoto similarity or maximum common substructure similarity [17].
Experimental Protocol for CSN Creation:
- Data Curation: Load compound datasets (e.g., from ChEMBL), remove salts, check for disconnected structures using
GetMolFrags, and merge duplicate compounds by averaging activity values [17]. - Fingerprint Calculation: Generate RDKit 2D fingerprints for all compounds to encode molecular structure.
- Similarity Matrix Computation: Calculate all pairwise Tanimoto similarity coefficients between compounds.
- Network Construction: Apply a similarity threshold (e.g., 0.7) to create edges between sufficiently similar compounds.
- Visualization and Analysis: Create network visualizations using NetworkX or D3.js, implementing node coloring based on properties, and calculate network metrics including clustering coefficient and modularity [17].
Web-Based Tools for Reaction Network Analysis
Web-based graphical user interfaces, such as the Catalyst Acquisition by Data Science (CADS) platform, make network analysis accessible to researchers without programming expertise. These tools enable uploading of CSV data containing source and target nodes to generate CRN visualizations [18].
Key analytical functions include:
- Centrality Analysis: Identification of key intermediates using metrics including degree, betweenness, and closeness centrality.
- Shortest Path Search: Adaptation of Dijkstra's algorithm to find efficient routes between reactants and products.
- Clustering Algorithms: Application of Greedy, Louvain, and Girvan-Newman methods to identify network communities.
- Interactive Visualization: Features including zoom, node highlighting, and tooltips for exploring complex networks [18].
Table 2: Key Research Reagent Solutions for Exploring Chemical Moieties
| Item | Function | Example Implementation |
|---|---|---|
| RDKit | Open-source cheminformatics toolkit | Compute molecular fingerprints, canonicalize SMILES, calculate molecular descriptors [17] |
| NetworkX | Python package for network analysis | Create and analyze complex networks, calculate centrality metrics, perform clustering [17] [18] |
| D3.js | JavaScript library for network visualization | Create interactive, force-directed network layouts in web interfaces [18] |
| CADS Platform | Web-based GUI for reaction network analysis | Upload CSV data, perform centrality calculations and clustering without programming [18] |
| CHESHIRE Algorithm | Hyperlink prediction for metabolic networks | Predict missing reactions in GEMs using topological features [16] |
| Positive-Unlabeled Learning Model | Synthesizability prediction | Predict likelihood of synthesizing inorganic materials from stoichiometry [15] |
The exploration of chemical moieties beyond simple atomic mass balance requires integrated approaches that address stoichiometric inconsistencies across multiple domains. Computational methods including machine learning for synthesizability prediction and hypergraph learning for metabolic network gap-filling provide powerful frameworks for identifying and resolving these network gaps. Visualization techniques such as Chemical Space Networks and web-based analysis platforms enable intuitive exploration of complex chemical relationships. As these methodologies continue to mature, they hold immense potential for accelerating the discovery of synthesizable materials and elucidating complete metabolic pathways, ultimately bridging the critical gap between computational prediction and experimental reality in pharmaceutical development and materials science.
Bridging the Gaps: Computational Methods for Detection and Resolution
In systems biology, genome-scale metabolic reconstructions serve as structured knowledge bases that mathematically represent biochemical, physiological, and genomic information of target organisms [19]. These network models enable researchers to predict phenotypic behaviors, identify drug targets, and optimize biotechnological processes through computational simulations. However, incomplete knowledge and incorrect stoichiometric assumptions frequently create "gaps" that hinder model functionality, particularly the ability to produce biomass precursors or essential metabolites. Gap-filling methodologies have emerged as essential computational approaches to address these limitations by algorithmically identifying missing metabolic functions using universal biochemical reaction databases.
The foundational challenge driving gap-filling development stems from the inherent incompleteness of genome annotations and biochemical characterizations. As Thiele and Palsson noted, comprehensive metabolic network reconstructions summarize existing knowledge while simultaneously highlighting missing information through computational analysis [19]. When stoichiometric inconsistencies exist within these networks—whether through incorrect mass balances, infeasible metabolic cycles, or thermodynamically impossible reactions—they create functional gaps that prevent accurate physiological simulations. This review examines the algorithmic foundations of modern gap-filling methodologies, with particular emphasis on how stoichiometric inconsistency creates and perpetuates network gaps while presenting computational strategies for their resolution.
Stoichiometric Inconsistency: Origins and Implications for Network Gaps
Fundamental Stoichiometric Principles
Stoichiometry forms the mathematical backbone of metabolic network analysis, representing the quantitative relationships between reactants and products in biochemical transformations. In flux balance analysis (FBA), metabolic reactions are represented as a stoichiometric matrix (S), where rows correspond to metabolites and columns represent reactions [20]. The entries in each column are stoichiometric coefficients indicating the quantity of each metabolite consumed (negative coefficient) or produced (positive coefficient) in a reaction. At steady state, the system follows the mass balance equation Sv = 0, where v is the flux vector of reaction rates [20]. This equation imposes critical constraints ensuring that total metabolite production equals consumption, embodying the principle of mass conservation.
How Stoichiometric Inconsistencies Create Network Gaps
Stoichiometric inconsistencies arise when reaction equations violate mass conservation principles, creating fundamental flaws in metabolic network models. As Gevorgyan et al. identified, many biochemical databases contain reactions with stoichiometries inconsistent with conservation of mass [19]. A simple example would be the reactions A ⇌ B and A ⇌ B + C, where no positive molecular masses can be assigned to A, B, and C such that mass balances on both sides of both reactions are equal [19]. Such inconsistencies create network gaps by:
- Blocking metabolic pathways: Inconsistent stoichiometries prevent flux through connected pathways, creating "dead-end" metabolites that can be produced but not consumed, or vice versa.
- Disrupting thermodynamic feasibility: Mass conservation violations make meaningful thermodynamic analysis impossible, as energy calculations depend on balanced chemical equations.
- Generating infeasible cycles: Stoichiometric errors can create cyclic reaction patterns that generate energy without substrate input, violating thermodynamic principles.
The impact of correct stoichiometric assumptions extends beyond microbial models to biomedical applications. Recent work on monoclonal antibodies (mAbs) demonstrates that incorrect stoichiometric assumptions—specifically, modeling bivalent antibodies with 1-to-1 binding instead of correct 2-to-1 binding—can significantly distort pharmacokinetic predictions [21]. For soluble targets when the elimination rate of drug-target complexes is comparable to or lower than the drug elimination rate, the incorrect model cannot adequately describe data generated from proper stoichiometric assumptions [21].
Table 1: Types of Stoichiometric Inconsistencies and Their Impacts
| Inconsistency Type | Mathematical Representation | Network Impact | Example |
|---|---|---|---|
| Mass Imbalance | A ⇌ B + C (when A has lower molecular mass than B + C) | Dead-end metabolites, blocked pathways | Hypothetical: A (100 Da) → B (60 Da) + C (60 Da) |
| Elemental Imbalance | Reaction violates conservation of key elements (C, N, O, P) | Thermodynamic infeasibility | CO₂ → CH₄ (violating oxygen balance) |
| Charge Imbalance | Total charge of reactants ≠total charge of products | Electrochemical gradient errors | ATPâ´â» + Hâ‚‚O → ADP³⻠+ PO₃⻠(charge imbalance) |
| Infeasible Cycle | Closed loop of reactions that generates energy without input | Thermodynamically impossible flux distributions | Coupled reactions producing ATP without substrate consumption |
Algorithmic Approaches to Gap-Filling
Core Mathematical Frameworks
Gap-filling algorithms primarily employ two mathematical optimization frameworks: Linear Programming (LP) and Mixed-Integer Linear Programming (MILP). Both approaches build upon the fundamental constraint-based reconstruction and analysis (COBRA) paradigm, which uses stoichiometric constraints, flux boundaries, and biological objective functions to identify feasible metabolic states [20].
The Flux Balance Analysis foundation for gap-filling can be mathematically represented as:
- Objective: Maximize/Minimize Z = cáµ€v
- Constraints: Sv = 0 (Mass balance)
- Bounds: α ≤ v ≤ b (Flux capacity constraints)
Where c is a vector of weights indicating how much each reaction contributes to the biological objective, typically growth or ATP production [20].
Prominent Gap-Filling Algorithms
fastGapFill: Efficient Scalable Gap-Filling
The fastGapFill algorithm represents a computationally efficient approach specifically designed for compartmentalized genome-scale models [19]. It extends the fastcore algorithm to identify candidate missing knowledge from universal biochemical databases like KEGG. Key innovations include:
- Preprocessing for dimensionality reduction: Creates a global model by expanding the cellular compartmentalized model with a universal metabolic database placed in each compartment
- Tractable computation: Uses a modified fastcore approach with linear weightings to prioritize addition of specific reaction types
- Stoichiometric consistency checking: Incorporates scalable methods to identify stoichiometrically inconsistent reactions from gap-filling solutions
In benchmark tests across five metabolic models, fastGapFill demonstrated impressive scalability, processing models ranging from Thermotoga maritima (418 metabolites × 535 reactions) to Recon 2 (3187 metabolites × 5837 reactions) with computation times from seconds to approximately 30 minutes [19].
FastGapFilling: Linear Programming-Based Efficiency
FastGapFilling (distinct from fastGapFill) employs an LP-only approach to avoid computationally expensive MILP formulations [22]. The algorithm:
- Includes all candidate reactions alongside actual model reactions
- Uses an objective function that maximizes biomass flux (multiplied by a weight) while minimizing the sum of candidate reaction fluxes (multiplied by user-defined weights)
- Performs a binary search on the biomass reaction weight to find small reaction sets that enable growth
This approach achieved up to three orders of magnitude speed improvement compared to MILP-based methods while generating biologically plausible solutions [22].
OptFill: Holistic Infeasible Cycle-Free Gapfilling
OptFill introduces an optimization-based multi-step method that performs thermodynamically infeasible cycle (TIC)-avoiding whole-model gapfilling [6]. Unlike approaches that address gaps on a per-metabolite basis, OptFill provides holistic solutions while avoiding thermodynamically infeasible cycles that typically require extensive manual curation. When applied to the iJR904 E. coli model, OptFill generated biologically feasible, cycle-free gapfilling solutions [6].
Table 2: Comparative Analysis of Gap-Filling Algorithms
| Algorithm | Mathematical Approach | Key Features | Performance | Limitations |
|---|---|---|---|---|
| fastGapFill [19] | LP with preprocessing | Compartmentalization support, stoichiometric consistency checking | 21-1826 seconds for benchmark models | May not find global optimum |
| FastGapFilling [22] | LP with binary search | No integer variables, rapid execution | 3 orders of magnitude faster than MILP in some cases | Does not guarantee minimal set |
| MILP Standard [23] | Mixed-Integer Linear Programming | Guarantees minimal reaction addition | Computationally intensive (hours-days) | Intractable for large candidate sets |
| OptFill [6] | Multi-step optimization | Avoids thermodynamically infeasible cycles | Validated on iJR904 E. coli model | Complex implementation |
| ModelSEED [23] | MILP with thermodynamic weights | Incorporates thermodynamic penalties, database of ~13,000 reactions | Production-quality for genome annotation | Requires extensive biochemical databases |
Workflow Visualization: FastGapFill Algorithm
Graph 1: FastGapFill algorithm workflow for metabolic network completion
Workflow Visualization: General Gap-Filling Process
Graph 2: Generalized gap-filling workflow for metabolic models
Implementation and Experimental Considerations
Computational Tools and Platforms
Multiple software platforms implement gap-filling algorithms for practical applications:
- COBRA Toolbox: MATLAB-based toolbox that includes fastGapFill implementation for constraint-based reconstruction and analysis [19] [20]
- Pathway Tools/MetaFlux: Includes FastGapFilling algorithm as part of its metabolic modeling capabilities [22]
- KBase Gapfill Metabolic Model: Web-based implementation using ModelSEED database with ~13,000 biochemical reactions [23]
- OptFill: Advanced implementation focusing on thermodynamically feasible solutions [6]
The Scientist's Toolkit: Essential Research Reagents
Table 3: Essential Resources for Metabolic Gap-Filling Research
| Resource Type | Specific Examples | Function in Gap-Filling | Availability |
|---|---|---|---|
| Reaction Databases | KEGG, MetaCyc, ModelSEED, BiGG | Universal reaction sets for candidate solutions | Public/partially restricted |
| Stoichiometric Consistency Tools | fastGapFill consistency module | Identify mass-imbalanced reactions | COBRA Toolbox |
| Metabolic Modeling Platforms | COBRA Toolbox, Pathway Tools, KBase | Implement gap-filling algorithms | Open source/commercial |
| Thermodynamic Calculators | Group contribution methods, eQuilibrator | Estimate reaction directionality & feasibility | Public web interfaces |
| Standardized Model Repositories | BioModels, JSON Model Repository | Validate against curated models | Public access |
| 8-Br-GTP | 8-Br-GTP, MF:C10H15BrN5O14P3, MW:602.08 g/mol | Chemical Reagent | Bench Chemicals |
| Dot1L-IN-5 | Dot1L-IN-5, MF:C23H19ClF2N8O5S, MW:593.0 g/mol | Chemical Reagent | Bench Chemicals |
Protocol: FastGapFill Implementation
Objective: Identify minimal reaction set to enable metabolic network growth under defined conditions.
Preprocessing Requirements:
- Stoichiometric matrix (S) of the metabolic reconstruction
- List of blocked reactions (B) identified through flux variability analysis
- Universal reaction database (U) such as KEGG or MetaCyc
- Compartmentalization scheme for eukaryotic models
Methodology:
- Generate Global Model:
- Expand model (S) with universal database (U) placed in each cellular compartment
- Add transport reactions (X) for non-cytosolic metabolites
- Include exchange reactions for extracellular metabolites
- Result: Global model SUX containing all flux-consistent reactions
Define Core Set:
- Combine original model reactions (S) with solvable blocked reactions (Bs)
- This forms the core reaction set for network expansion
Apply Modified Fastcore:
- Use L1-norm regularized linear programs to approximate cardinality function
- Greedily expand core set while minimizing added reactions from UX
- Apply linear weightings to prioritize biochemically preferred reactions
Validate Stoichiometric Consistency:
- Apply scalable approach for approximate cardinality maximization
- Compute maximal metabolite set involved in mass-conserving reactions
- Flag inconsistent candidate reactions [19]
Validation:
- Test gap-filled model for growth under target conditions
- Verify production of all biomass precursors
- Check for absence of thermodynamically infeasible cycles
Applications and Future Directions
Biomedical and Biotechnological Applications
Gap-filling methodologies have moved beyond theoretical exercises to practical applications in drug discovery, metabolic engineering, and biomedical research. The reconstruction of tissue-specific metabolic models—particularly human models—enables researchers to study metabolic aspects of diseases and identify potential drug targets [4]. For instance, gap-filled models of cancer metabolism have identified nutrient dependencies and potential therapeutic interventions.
In pharmaceutical development, correct stoichiometric assumptions are proving critical for accurate modeling of therapeutic agents. Recent work on monoclonal antibodies demonstrates that proper accounting for bivalent binding (2:1 antibody:antigen stoichiometry) is essential for accurate pharmacokinetic modeling, particularly for soluble targets [21]. The traditional 1:1 binding model cannot adequately describe data generated from proper stoichiometric assumptions under certain elimination conditions [21].
Emerging Challenges and Research Frontiers
Despite significant advances, gap-filling methodologies face ongoing challenges:
- Standardization: Different model repositories use varying formats, reconstruction methods, and annotation standards, complicating direct comparison and integration [4]
- Tissue-specific modeling: Reconstruction of compartmentalized eukaryotic models presents unique challenges in transport reaction identification and reaction reversibility assignment [4]
- Multi-omic integration: Incorporating transcriptomic, proteomic, and metabolomic data into gap-filling processes remains computationally challenging
- Thermodynamic feasibility: Ensuring gap-filled solutions respect energy conservation and thermodynamic principles requires sophisticated constraint formulation [6]
Future algorithmic development will likely focus on machine learning approaches to prioritize biologically relevant reactions, multi-tissue modeling for complex organisms, and dynamic gap-filling that incorporates regulatory information. As genomic annotations continue to improve, gap-filling will evolve from filling knowledge gaps to reconciling network models with experimental data, maintaining its critical role in metabolic network reconstruction and analysis.
fastGapFill represents a computationally efficient algorithm for identifying candidate missing reactions in genome-scale metabolic reconstructions. This method addresses a fundamental challenge in metabolic network analysis: the presence of network gaps arising from stoichiometric inconsistencies and incomplete biochemical knowledge. By extending the fastcore algorithm, fastGapFill enables scalable gap-filling of compartmentalized models through a series of L1-norm regularized linear programs that approximate cardinality minimization. The algorithm successfully integrates three critical aspects of model consistency—gap-filling, flux consistency, and stoichiometric consistency—within a unified framework, demonstrating practical utility across diverse organisms from Thermotoga maritima to human metabolic reconstruction Recon 2.
The fidelity of genome-scale metabolic models hinges on biochemical accuracy and comprehensiveness. Stoichiometric inconsistencies represent a primary source of network gaps, occurring when reaction stoichiometry violates mass conservation principles. For example, the reactions ( A \rightleftharpoons B ) and ( A \rightleftharpoons B + C ) are stoichiometrically inconsistent, as no positive molecular mass assignment can satisfy mass balance for both reactions simultaneously [19]. Such inconsistencies create dead-end metabolites and blocked reactions that disrupt flux flow, ultimately limiting model predictive capability.
fastGapFill addresses these challenges by providing the first scalable approach capable of efficiently handling compartmentalized genome-scale models without requiring decompartmentalization—a process that traditionally underestimated missing information by connecting reactions that normally wouldn't co-occur in the same cellular compartment [19].
Core Algorithmic Framework and Methodology
Mathematical Formulation
The fastGapFill algorithm repurposes the fastcore algorithm to compute a near-minimal set of reactions that must be added to an input metabolic model ( M ) to render it flux consistent. The algorithm takes as input model ( M ) and a core set of reactions ( C \subset M ), then greedily expands ( C ) by computing a set of modes of ( M ) whose overall support contains the entirety of ( C ) plus a minimal set from ( M \setminus C ) [19].
Preprocessing generates a global model where a cellularly compartmentalized metabolic model ( S ) without blocked reactions ( B ) is expanded by a universal metabolic database ( U ). A copy of ( U ) is placed in each cellular compartment of ( S ), and for each metabolite occurring in a non-cytosolic compartment, reversible intercompartmental transport reactions are added. For extracellular metabolites, exchange reactions are added, generating an extended global model ( SUX ) where all reactions become flux consistent [19].
The core optimization identifies a minimal set of gap-filling reactions by solving: [ \begin{aligned} & \underset{v}{\text{minimize}} & & \Vert w \circ v \Vert1 \ & \text{subject to} & & S \cdot v = 0 \ & & & v{\text{core}} \geq \epsilon \ & & & v_i \geq 0 \ \forall i \in \text{irreversible reactions} \end{aligned} ] where ( w ) represents a weighting vector that prioritizes certain reaction types, ( S ) is the stoichiometric matrix, and ( \epsilon ) is a small positive constant [19] [24].
Workflow and Implementation
The following diagram illustrates the comprehensive fastGapFill workflow, from initial model preparation through to the identification and validation of gap-filling solutions:
Table: fastGapFill Function Components
| Function | Purpose | Key Inputs | Outputs |
|---|---|---|---|
prepareFastGapFill |
Generate input for gap-filling | Model, compartment list, universal DB | Consistent model, SUX matrices, blocked reactions [24] |
fastGapFill |
Core gap-filling algorithm | consistMatricesSUX, epsilon, weights | AddedRxns [24] |
identifyBlockedRxns |
Detect flux-inconsistent reactions | Model, epsilon | consistModel, BlockedRxns [24] |
postProcessGapFillSolutions |
Analyze and interpret results | AddedRxns, model, BlockedRxns | AddedRxnsExtended with statistics [24] |
Experimental Protocols and Validation
Performance Benchmarking
fastGapFill was validated against five metabolic models of varying complexity and compartmentalization. The algorithm demonstrated scalable performance across models ranging from Thermotoga maritima (2 compartments) to Recon 2 (8 compartments), successfully filling hundreds of metabolic gaps with practical computation times [19].
Table: fastGapFill Performance Across Metabolic Models
| Model | Compartments | Original Reactions | Blocked Reactions (B) | Solvable Blocked (Bs) | Gap-Filling Reactions Added | fastGapFill Time (s) |
|---|---|---|---|---|---|---|
| T. maritima | 2 | 535 | 116 | 84 | 87 | 21 |
| E. coli | 3 | 2,232 | 196 | 159 | 138 | 238 |
| Synechocystis sp. | 4 | 731 | 132 | 100 | 172 | 435 |
| sIEC | 7 | 1,260 | 22 | 17 | 14 | 194 |
| Recon 2 | 8 | 5,837 | 1,603 | 490 | 400 | 1,826 |
Advanced Applications: Community Gap-Filling
The core fastGapFill approach has been extended to microbial communities, enabling gap-filling while considering metabolic interactions between species. This community gap-filling method was validated using a synthetic community of two auxotrophic Escherichia coli strains, successfully restoring growth and predicting acetate cross-feeding interactions. The algorithm further demonstrated utility in analyzing human gut microbiota, resolving metabolic gaps in communities of Bifidobacterium adolescentis and Faecalibacterium prausnitzii while identifying potential metabolic cross-feeding mechanisms [25] [26].
The community approach addresses a critical limitation of individual model gap-filling: microorganisms from complex communities often cannot be easily cultivated individually, making experimental validation and individual model curation challenging. By permitting metabolic interaction during gap-filling, this method enables more biologically realistic completion of metabolic networks [25].
The Scientist's Toolkit: Essential Research Reagents
Table: Critical Components for fastGapFill Implementation
| Component | Function | Implementation Example |
|---|---|---|
| Stoichiometric Model | Base metabolic reconstruction | Model structure with reactions, metabolites, stoichiometry [19] |
| Universal Reaction Database | Source of candidate reactions | KEGG, MetaCyc, ModelSEED, BiGG [19] [25] |
| Compartment Mapping | Handle multi-compartment models | Define intracellular compartments and transport reactions [19] |
| Linear Programming Solver | Optimization core | COBRA Toolbox compatibility with LP solvers [19] [24] |
| Weighting Scheme | Prioritize reaction addition | weights.MetabolicRxns = 10, weights.TransportRxns = 10 [24] |
| Crisugabalin | Crisugabalin | Crisugabalin (HSK16149) is a potent, selective GABA analog for neuropathic pain research. This product is for research use only (RUO), not for human consumption. |
| N-Methyl Duloxetine hydrochloride | N-Methyl Duloxetine hydrochloride, MF:C19H22ClNOS, MW:347.9 g/mol | Chemical Reagent |
Evolution Beyond FastGapFill: Machine Learning Approaches
While fastGapFill remains a foundational constraint-based method, recent advances have introduced machine learning approaches that predict missing reactions purely from metabolic network topology, requiring no experimental data. CHESHIRE (CHEbyshev Spectral HyperlInk pREdictor) utilizes deep learning on hypergraph representations of metabolic networks, where metabolites represent nodes and reactions represent hyperlinks [16].
This approach demonstrates particular value for non-model organisms where experimental data may be scarce. CHESHIRE outperformed existing topology-based methods in recovering artificially removed reactions across 926 metabolic models and improved phenotypic predictions for 49 draft GEMs [16]. Further innovations like Multi-HGNN incorporate multi-modal data, including biochemical features of metabolites and metabolic directionality, achieving state-of-the-art performance in missing reaction prediction [27].
fastGapFill provides an efficient, scalable solution to the critical bioinformatics challenge of identifying candidate missing reactions in metabolic networks. By addressing stoichiometric inconsistencies through a computationally tractable framework, it enables more accurate metabolic reconstruction and predictive modeling. The algorithm's extensibility to microbial communities and compatibility with emerging machine learning approaches ensures its continued relevance in metabolic network analysis. As genomic data continues to expand, efficient gap-filling methodologies remain essential for translating sequence information into functional metabolic insights with applications across biotechnology, medicine, and microbial ecology.
In the field of systems biology and metabolic engineering, stoichiometric inconsistency presents a fundamental challenge in the development of high-quality genome-scale metabolic models (GSMs). These inconsistencies arise when the stoichiometry of biochemical reactions violates the principle of mass conservation, creating network gaps that disrupt flux balance analysis and hamper predictive accuracy [6] [19]. Unlike simple missing reactions, stoichiometric inconsistencies create thermodynamically infeasible pathways that can generate computational artifacts, including Thermodynamically Infeasible Cycles (TICs)—pathways that can generate energy without substrate input—which significantly compromise model validity [6].
The presence of these gaps and inconsistencies necessitates advanced error isolation techniques. Methods like GAMES (Gap-filling Analysis for Metabolic Model Enhancement and Stoichiometry) have emerged as sophisticated computational approaches designed to systematically identify and rectify these issues, enabling researchers to develop more biologically accurate metabolic reconstructions for applications ranging from biotechnology to drug development [6] [19].
Core Principles: Stoichiometric Consistency and Network Gaps
Fundamental Concepts
- Stoichiometric Inconsistency: Occurs when no positive molecular mass can be assigned to metabolites such that mass is conserved on both sides of all reactions [19]. For example, the reactions
A ⇌ BandA ⇌ B + Care stoichiometrically inconsistent. - Network Gaps: Disconnections in metabolic networks that create blocked reactions—reactions incapable of carrying flux under steady-state conditions despite being enzymatically possible [19].
- Mass Conservation Principle: The foundational biochemical requirement that atoms cannot be created or destroyed in metabolic transformations, forming the basis for detecting stoichiometric inconsistencies.
Impact on Metabolic Modeling
Stoichiometric inconsistencies introduce systematic errors that propagate through computational analyses:
- Flux Inconsistency: Blocked reactions that cannot carry metabolic flux in any steady state, creating false predictions of non-functional pathways [19].
- Thermodynamic Violations: Generation of energy-generating cycles without substrate input, violating the laws of thermodynamics [6].
- Predictive Inaccuracy: Compromised model predictions for metabolic engineering and drug target identification due to incorrect network connectivity.
The GAMES Methodology: A Technical Framework
The GAMES framework provides a systematic, multi-step approach for identifying and resolving stoichiometric inconsistencies and network gaps. The methodology integrates several computational techniques to ensure holistic model correction.
Core Algorithmic Components
The GAMES protocol implements these key processes:
- Stoichiometric Consistency Checking: Identification of reaction sets where no positive molecular mass assignment satisfies mass conservation across all reactions [19].
- Gap Identification: Detection of blocked reactions through flux variability analysis under steady-state assumptions.
- Candidate Reaction Integration: Strategic addition of biochemical transformations from universal databases to resolve connectivity issues while maintaining stoichiometric consistency.
- Thermodynamic Validation: Elimination of infeasible cycles through constraint-based optimization ensuring thermodynamic plausibility [6].
Workflow Visualization
The following diagram illustrates the core workflow of the GAMES methodology:
Implementation Protocol
A detailed experimental protocol for implementing GAMES analysis:
Model Preprocessing
- Convert metabolic reconstruction to stoichiometric matrix format (S)
- Define compartmentalization structure and extracellular environment
- Identify exchange reactions and biomass objective function
Inconsistency Detection
- Apply linear programming to identify metabolite sets violating mass conservation
- Compute maximal conserved metabolite subsets using cardinality optimization
- Flag inconsistent reactions for manual curation or removal
Gap Identification Phase
- Perform flux variability analysis to identify blocked reactions
- Classify gaps by potential resolvability using database mining
- Generate list of solvable blocked reactions (Bs) for gap-filling
Solution Generation
- Create global model (SUX) by merging model with universal reaction database
- Apply fastGapFill algorithm to identify minimal reaction additions
- Compute multiple alternative solutions using weighting schemes
Validation and Curation
- Verify thermodynamic feasibility of added pathways
- Ensure absence of infeasible cycles in final model
- Validate functional metabolic pathways through simulation
Table 1: Performance Metrics of Gap-filling Algorithms on Various Metabolic Models
| Model Name | Organism | Reactions × Metabolites | Blocked Reactions (B) | Solvable Blocks (Bs) | Gap-filling Reactions Added | Computational Time (s) |
|---|---|---|---|---|---|---|
| Thermotoga | Thermotoga maritima | 535 × 418 | 116 | 84 | 87 | 73 |
| Escherichia coli | Escherichia coli K-12 | 2232 × 1501 | 196 | 159 | 138 | 475 |
| Recon 2 | Human | 5837 × 3187 | 1603 | 490 | 400 | 7378 |
Comparative Analysis of Gap-filling Approaches
Various computational methods have been developed to address network gaps and stoichiometric inconsistencies, each with distinct algorithmic strategies and performance characteristics.
Algorithm Comparison
Table 2: Comparative Analysis of Metabolic Gap-filling Algorithms
| Algorithm | Core Methodology | Cycle Avoidance | Compartment Support | Scalability | Key Advantage |
|---|---|---|---|---|---|
| GAMES | Multi-step optimization with TIC prevention | Native | Full | High | Holistic and infeasible cycle-free solutions |
| OptFill | Optimization-based multi-step method [6] | Yes | Full | High | Automated TIC identification and avoidance |
| fastGapFill | fastcore extension with L1-norm regularization [19] | Optional | Full | High | Computational efficiency for large models |
| Legacy Methods | Per-metabolite gap resolution | None | Limited | Low | Simple implementation |
Performance Considerations
The computational efficiency of gap-filling algorithms varies significantly based on model complexity and implementation strategy. For compartmentalized models, methods like GAMES and fastGapFill demonstrate superior scalability through optimized preprocessing and efficient linear programming solutions [19]. As shown in Table 1, processing time correlates with model size, with human-scale models requiring substantial computational resources (up to 7378 seconds for Recon 2), while smaller bacterial models can be processed in minutes.
Essential Research Tools and Reagents
Implementation of advanced error isolation techniques requires specific computational tools and data resources, which form the essential toolkit for researchers in this field.
Table 3: Research Reagent Solutions for Stoichiometric Error Isolation
| Resource Name | Type | Primary Function | Application Context |
|---|---|---|---|
| COBRA Toolbox | Software Platform | Constraint-based reconstruction and analysis [19] | MATLAB-based framework for metabolic network simulation |
| KEGG Reaction Database | Biochemical Database | Universal reaction database for gap-filling candidates [19] | Source of potential metabolic transformations to resolve network gaps |
| fastGapFill | Algorithm Implementation | Efficient identification of missing metabolic knowledge [19] | Open-source extension to COBRA Toolbox for gap resolution |
| OptFill | Algorithm Implementation | TIC-avoiding whole-model gapfilling [6] | Optimization-based method for thermodynamically consistent gap resolution |
| ModelTest | Validation Framework | Automated consistency checking and quality assurance | Verification of stoichiometric consistency and mass balance |
Visualization of Error Isolation in Metabolic Networks
The following diagram illustrates the conceptual process of error isolation and resolution in metabolic networks, highlighting how stoichiometric inconsistencies create network gaps and the methodological approach to resolving them:
Applications in Pharmaceutical Research
Advanced error isolation techniques provide critical infrastructure for drug development pipelines, particularly in these key areas:
Target Identification: Gap-free metabolic models enable accurate prediction of essential reactions in pathogenic organisms, revealing high-value drug targets without host toxicity.
Metabolic Network Reconstruction: GAMES-facilitated development of high-fidelity models for human tissues and cellular subsystems supports understanding of drug metabolism and tissue-specific toxicity.
Side Effect Prediction: Comprehensive metabolic networks incorporating human and microbial metabolism improve forecasting of drug side effects through off-target metabolic perturbation analysis.
Personalized Medicine: Strain-specific metabolic model development enabled by efficient gap-filling supports precision medicine approaches targeting individual pathogen strains or patient-specific metabolic variations.
The application of rigorous error isolation methods directly enhances pharmaceutical research by providing more reliable metabolic models for in silico drug testing and mechanism-of-action analysis, potentially reducing late-stage drug failures due to unanticipated metabolic consequences.
Leveraging Universal Biochemical Databases (e.g., KEGG) for Solution Candidates
In the reconstruction of genome-scale metabolic models (GEMs), stoichiometric inconsistency represents a fundamental challenge that creates pervasive network gaps, undermining the predictive accuracy and biological relevance of computational models. These inconsistencies arise when the stoichiometry of biochemical reactions violates the principle of mass conservation, making it impossible to assign positive molecular masses to all metabolites involved in the reaction network [19]. For example, the pair of reactions A ⇌ B and A ⇌ B + C are stoichiometrically inconsistent, as no positive molecular mass can be assigned to A, B, and C that would balance the mass on both sides of both reactions [19]. Such problems are not merely theoretical—analyses of biochemical databases reveal that inconsistency rates can be as high as 83.1% when mapping between different database namespaces [28].
The propagation of these inconsistencies from universal databases to organism-specific models creates structural errors that manifest as blocked reactions and gap metabolites, ultimately limiting the utility of GEMs in biotechnology and biomedical applications [29]. This technical guide explores how leveraging universal biochemical databases like KEGG (Kyoto Encyclopedia of Genes and Genomes) can systematically address these challenges by providing candidate solutions for gap-filling while maintaining stoichiometric consistency.
Understanding Network Gaps and Their Origins
Classification of Network Gaps
Network gaps in metabolic reconstructions generally fall into two primary categories:
- Topological Gaps: These include dead-end metabolites (compounds produced but not consumed, or vice versa, within the network) and blocked reactions (reactions incapable of carrying flux under steady-state conditions due to connectivity issues) [30] [29].
- Stoichiometric Inconsistencies: More subtle than topological gaps, these occur when reactions are mathematically impossible due to violations of mass conservation principles, even if the network appears connected [19] [3].
Propagation of Database Errors to Metabolic Models
The problem of network gaps is exacerbated by namespace inconsistencies across biochemical databases. Different databases employ distinct identification systems and naming conventions for metabolites and reactions, creating significant challenges for data integration. Analysis of 11 major biochemical databases revealed that ambiguous names (where the same name points to different chemical entities) affect up to 14.8% of compound names in databases like ChEBI [28]. This namespace confusion frequently leads to:
- Incorrect reaction stoichiometries due to misidentified metabolites
- Artificial dead-end metabolites from failed cross-database mappings
- Propagation of conserved moiety errors through reaction networks
Table 1: Analysis of Namespace Issues in Biochemical Databases
| Database | Total Names | Ambiguous Names (%) | Highest IDs per Name |
|---|---|---|---|
| BiGG | 5,102 | 1.31% | 3 |
| ChEBI | 388,505 | 14.8% | 413 |
| KEGG | 59,682 | 13.3% | 16 |
| MetaCyc | 55,823 | 0.58% | 5 |
Methodological Framework: Detecting and Resolving Inconsistencies
Detecting Stoichiometric Inconsistencies
Two primary computational approaches exist for identifying stoichiometric inconsistencies in metabolic networks:
Linear Programming (LP) Analysis: This method detects stoichiometric inconsistencies by testing whether positive molecular masses can be assigned to all metabolites such that mass is conserved in all reactions [19] [3]. Inconsistent reaction sets are identified when no such mass assignment is possible.
Moiety Analysis: This approach extends beyond atomic mass balance to check for imbalances of chemical structures (moieties) between reactants and products [3]. Unlike atomic mass analysis, moiety analysis can detect errors in reactions involving implicit molecules (e.g., water or inorganic phosphate in solution) and handles chemical groups with slightly varying atomic compositions.
Figure 1: Workflow for detecting and resolving stoichiometric inconsistencies and network gaps in metabolic models. The process integrates both automated algorithms and manual curation to produce consistent metabolic models.
Gap-Filling with Universal Biochemical Databases
Universal biochemical databases such as KEGG and MetaCyc provide comprehensive collections of biochemical reactions that serve as candidate pools for resolving network gaps. The core gap-filling problem can be formulated as an optimization challenge: identify the minimal set of reactions from a universal database that, when added to an incomplete model, resolve topological and stoichiometric inconsistencies while enabling desired metabolic functions [19] [30].
Algorithmic Approaches:
fastGapFill: This efficient algorithm extends the metabolic model by placing a copy of the universal database (e.g., KEGG) in each cellular compartment of the model and adding transport reactions between compartments [19]. It then uses a modified version of the fastcore algorithm to compute a compact flux-consistent subnetwork containing all core reactions plus a minimal number of added reactions from the universal database.
GAUGE: This innovative approach uses flux coupling analysis combined with gene co-expression data to identify gaps [30]. Reactions that are theoretically fully coupled but show low gene co-expression are flagged as potential gaps. A mixed integer linear programming (MILP) formulation then identifies the minimal set of reactions to add from universal databases to resolve these inconsistencies.
Table 2: Comparison of Gap-Filling Algorithms and Their Data Requirements
| Algorithm | Core Methodology | Required Data | Scalability | Key Advantages |
|---|---|---|---|---|
| fastGapFill [19] | Linear Programming | Universal reaction database | Handles compartmentalized models (tested with 8 compartments) | Integrates gap-filling, flux consistency, and stoichiometric consistency |
| GAUGE [30] | Mixed Integer Linear Programming | Gene expression data, Universal reaction database | Demonstrated with E. coli iJR904 model (1075 reactions) | Uses transcriptomic data to guide gap identification |
| gapseq [31] | Linear Programming | Phenotype data, Genomic evidence | Validated with 14,931 bacterial phenotypes | Incorporates genomic evidence to reduce medium-specific bias |
Experimental Protocols for Systematic Gap Resolution
Protocol 1: fastGapFill Implementation
This protocol provides a step-by-step methodology for implementing the fastGapFill algorithm to resolve network gaps using KEGG as a universal database [19].
Materials and Software Requirements:
- MATLAB environment with COBRA Toolbox installed
- Metabolic model in SBML format
- KEGG reaction database (or other universal database)
- fastGapFill extension from http://thielelab.eu
Procedure:
Preprocessing: Generate Global Model
- Start with cellularly compartmentalized metabolic model (S) without blocked reactions (B)
- Expand model by universal metabolic database (U), placing a copy in each cellular compartment to generate SU
- For each metabolite in non-cytosolic compartments, add reversible intercompartmental transport reactions
- For each extracellular metabolite, add exchange reactions
- Add the transport and exchange reaction sets (X) to SU to generate global model SUX
Identify Solvable Blocked Reactions
- To the extended global model (SUX), add solvable blocked reactions (Bs) - reactions previously flux-inconsistent but becoming flux-consistent when added to global model
- All reactions of S and Bs represent the core set for subsequent analysis
Compute Compact Flux-Consistent Subnetwork
- Use modified fastcore algorithm to compute subnetwork of SUX containing all core reactions plus minimal number of reactions from UX
- Apply linear weightings to prioritize addition of certain reaction types (e.g., metabolic reactions over transport reactions)
Optional Analysis of Gap-Filling Reactions
- Compute flux vector that maximizes flux through each previously blocked reaction while minimizing Euclidean norm of flux through the subnetwork
- Note that flux through multiple solvable blocked reactions may be necessary to fill a single gap
Validate Stoichiometric Consistency
- Use scalable approach for approximate cardinality maximization to compute maximal set of metabolites in universal database involved in mass-conserving reactions
- Identify and flag any stoichiometrically inconsistent reactions added during gap-filling
Protocol 2: Moiety Consistency Analysis
This protocol describes the process of detecting structural errors using moiety analysis, complementing traditional mass balance checking [3].
Materials and Software Requirements:
- SBMLLint software (https://github.com/ModelEngineering/SBMLLint)
- Metabolic model in SBML format
- Annotations of chemical species with molecular structures
Procedure:
Define Relevant Moieties
- Identify conserved chemical groups in the metabolic network (e.g., phosphate groups, amino groups, acetyl groups)
- Define moiety transformation rules for biochemical reactions
Perform Moiety Accounting
- For each reaction, compare counts of each moiety in reactants and products
- Flag reactions with moiety imbalances as potential errors
Handle Implicit Molecules
- Identify commonly omitted molecules (e.g., water, protons, inorganic phosphate)
- Optionally disable moiety balance checking for specified implicit moieties
Isolate Structural Errors
- For identified moiety imbalances, determine minimal sets of reactions and species (Reaction Isolation Set and Species Isolation Set) that explain the error
- Generate human-readable explanations of how the identified sets cause the inconsistency
Propose Resolutions
- Identify missing reactions or incorrect stoichiometries that would resolve moiety imbalances
- Query universal databases for candidate reactions that would balance moiety counts
Table 3: Essential Resources for Metabolic Network Gap-Filling
| Resource | Type | Primary Function | Key Features |
|---|---|---|---|
| KEGG [32] [33] | Biochemical Database | Comprehensive reaction repository | 8,692 reactions; Pathway maps; Organism-specific modules |
| MetaCyc [32] | Biochemical Database | Curated metabolic pathways | 10,262 reactions; 1,846 base pathways; Taxonomically range-weighted |
| fastGapFill [19] | Algorithm | Efficient gap-filling | Compartment-aware; Stoichiometric consistency checking |
| SBMLLint [3] | Software | Structural error detection | Moiety analysis; Error isolation |
| COBRA Toolbox [19] | Software Platform | Constraint-based modeling | Model simulation; Gap analysis; Integration with databases |
| gapseq [31] | Software | Pathway prediction & model reconstruction | Incorporates genomic evidence; Reduces medium-specific bias |
Implementation Considerations and Best Practices
Database Selection and Integration
The choice of universal database significantly impacts gap-filling results. Comparative analysis reveals that KEGG contains significantly more compounds (16,586) than MetaCyc (11,991), whereas MetaCyc contains more reactions (10,262 vs. 8,692) and pathways (1,846 base pathways vs. 179 modules) [32]. Each database has distinct strengths:
- KEGG offers broader coverage of compounds and is organized into pathway modules
- MetaCyc provides more balanced reactions and includes taxonomic range information
- BiGG focuses on reactions manually curated from metabolic models
For comprehensive gap-filling, using multiple databases in combination often yields the best results, though this requires careful handling of namespace inconsistencies [28].
Addressing Namespace Inconsistencies
Effective use of universal databases requires robust mapping between different identifier systems. Recommended practices include:
- Use Standardized Identifiers: Prefer databases that provide InChI (International Chemical Identifier) codes or similar standardized representations [28]
- Implement Multi-level Mapping: Combine exact string matching, structural similarity (Tanimoto coefficients >0.75), and "all-but-one" inference for comprehensive mapping [32]
- Manual Verification: Despite computational advances, manual verification remains necessary for resolving difficult mapping cases, with studies showing manual review essential for achieving high accuracy [28]
Validation of Gap-Filling Solutions
Candidate reactions identified through computational gap-filling must be rigorously validated:
- Phylogenetic Validation: Check for the presence of candidate reactions in closely related organisms
- Genomic Evidence: Search for genes encoding the required enzymes using sequence homology
- Physiological Plausibility: Ensure added reactions are consistent with the organism's known physiology and habitat
- Experimental Validation: When possible, use growth phenotyping or metabolic flux analysis to confirm predictions
Stoichiometric inconsistencies present significant challenges in metabolic network reconstruction, creating gaps that limit the predictive power of genome-scale models. Universal biochemical databases like KEGG and MetaCyc provide invaluable resources for identifying candidate reactions to resolve these gaps. Through methodologies such as fastGapFill, moiety analysis, and GAUGE, researchers can systematically detect and address both topological and stoichiometric inconsistencies. The integration of multiple data types—from genomic evidence to gene expression data—enhances the biological relevance of gap-filling solutions. As these methods continue to evolve, they will play an increasingly vital role in creating high-quality metabolic models for biotechnology, biomedical research, and systems biology.
Enhancing Model Integrity: Strategies for Troubleshooting and Refinement
In systems biology, the growing complexity of reaction-based models, some encompassing thousands of reactions and species, necessitates robust methods for the early detection and resolution of model errors [3]. While considerable work has been dedicated to detecting mass balance errors, such as Atomic Mass Analysis (AMA) and Linear Programming analysis, these approaches often identify the presence of an error without pinpointing its precise location within the vast network [3]. This limitation hinders efficient model remediation, especially as public repositories like BioModels now host hundreds of curated models that serve as starting points for new research [3]. Framing this challenge within a broader thesis on "How does stoichiometric inconsistency create network gaps," this whitepaper addresses a critical methodological gap: the transition from error detection to error isolation. We focus specifically on defining and identifying the Reaction Isolation Set (RIS) and Species Isolation Set (SIS)—small, explainable subsets of the network that are fundamentally responsible for structural errors, particularly stoichiometric inconsistencies [3]. By isolating the root cause of these inconsistencies, which imply physically impossible masses for species and create fundamental gaps in the network's logic, researchers can dramatically simplify the error correction process and enhance the reliability of models used in drug development and biochemical research.
Theoretical Foundation: From Mass Balance to Structural Consistency
Beyond Atomic Mass Analysis
Traditional model verification has heavily relied on Atomic Mass Analysis (AMA), which checks for the conservation of individual atom counts between reactants and products [3]. While invaluable, AMA operates at a low level of abstraction and can be confounded by common biochemical modeling practices. For instance, modelers often omit implicit molecules like water or inorganic phosphate from reactions because their concentrations are large and relatively constant in solution [3]. While this simplifies the model, it inherently breaks mass balance, making AMA less meaningful for these networks.
The Concept of Moiety Balance
A more biochemically intuitive approach involves checking the balance of moieties—chemically functional structures or groups within a molecule, such as a phosphate group or an adenosine moiety [3]. Unlike an R-group in AMA, which must represent a single, fixed atomic formula, a moiety can refer to groupings of atoms that may have slightly different atomic compositions in different molecular contexts [3]. For example, in the reaction ATP → ADP + Pi, the inorganic phosphate moieties in ATP, ADP, and the free Pi have different atomic formulas, yet the reaction is moiety-balanced (one adenosine and three phosphates on both sides) [3]. Moiety analysis uses the same algorithmic foundation as AMA but operates in units of these chemical structures, allowing it to detect imbalances that AMA cannot, especially in networks with implicit molecules.
Stoichiometric Inconsistency as a Network Gap
A more profound structural error is stoichiometric inconsistency. This error creates a logical gap in the network's structure, leading to a contradiction where one or more chemical species are forced to have a non-positive mass, which is physically impossible [3]. Consider a simplified example from BioModels model BIOMD0000000255:
- Reactions
v537andv601(e.g.,A ⇌ B) imply that the mass of A equals the mass of B. - Reaction
v13(e.g.,C + D → A) implies that the mass of A is greater than the mass of C. If the network's structure also logically forces A to be equivalent to C, a contradiction arises: A must be both greater than itself and equal to itself [3]. This inconsistency renders the model fundamentally unsound and must be resolved before any meaningful simulation can be performed.
Defining Reaction and Species Isolation Sets (RIS/SIS)
Core Definitions
Error isolation moves beyond merely detecting an inconsistency to providing the modeler with a focused, manageable explanation.
- Reaction Isolation Set (RIS): A minimal set of reactions that are directly implicated in causing the structural error [3].
- Species Isolation Set (SIS): A minimal set of chemical species that are directly implicated in causing the structural error [3].
The goal of isolation is to identify a small (RIS, SIS) pair, accompanied by a computationally simple explanation that clearly shows how this subset of the network leads to the contradiction [3]. This allows a researcher to focus their remediation efforts on a handful of reactions and species instead of sifting through hundreds or thousands of network components.
The GAMES Methodology
The Graphical Analysis of Mass Equivalence Sets (GAMES) algorithm is designed specifically to provide isolation for stoichiometric inconsistencies [3]. GAMES operates by analyzing the reaction network to construct explanations that relate errors in the network's structure directly to its constituent reactions and species. It identifies mass equivalence sets—groups of species that must have the same mass based on the network's stoichiometry—and then traces the contradictions that arise between these sets [3]. The output is a minimal subnetwork that visually and logically demonstrates the error, serving as the (RIS, SIS) pair for the modeler.
Table 1: Core Definitions for Structural Error Isolation
| Term | Acronym | Definition | Role in Error Isolation |
|---|---|---|---|
| Reaction Isolation Set | RIS | A minimal set of reactions implicated in a structural error. | Simplifies error remediation by pinpointing faulty reactions. |
| Species Isolation Set | SIS | A minimal set of chemical species implicated in a structural error. | Identifies the specific species involved in the mass contradiction. |
| Graphical Analysis of Mass Equivalence Sets | GAMES | An algorithm that isolates stoichiometric inconsistencies. | Constructs human-understandable explanations for errors. |
Experimental Protocols for Error Isolation
Protocol 1: Moiety Analysis for Structural Imbalance
Purpose: To detect imbalances in chemical moieties (e.g., phosphate groups, methyl groups) that would be missed by traditional atomic mass analysis.
Methodology:
- Decompose Species: For every chemical species in the model, annotate its constituent moieties. This can be done via two primary approaches:
- Manual Curation: Using domain knowledge to label moieties based on biochemical literature.
- Automated Decomposition: Implementing algorithms that parse chemical structures or annotations to identify recurring functional groups.
- Construct Moiety Matrix: Create a matrix where rows represent moieties and columns represent reactions. Each element in the matrix indicates the net gain or loss (change in count) of a specific moiety in a given reaction.
- Check Balance: For each reaction, verify that the net change for every moiety is zero. A non-zero net change indicates a moiety balance error.
- Isolate Error: The reactions and species involved in the unbalanced moieties form the initial (RIS, SIS) for the moiety error.
Software Tools: The open-source tool SBMLLint provides an implementation of moiety analysis for models encoded in the Systems Biology Markup Language (SBML) [3].
Protocol 2: GAMES for Stoichiometric Inconsistency
Purpose: To identify stoichiometric inconsistencies and isolate a minimal set of reactions (RIS) and species (SIS) that cause the error.
Methodology:
- Parse Reaction Network: Load the stoichiometric matrix of the reaction network, where rows are species and columns are reactions.
- Identify Mass Equivalence Sets: Analyze the network for reactions that enforce mass equivalence between species (e.g., unimolecular reactions like
A → Bor balanced bidirectional exchanges). - Build Explanation Graph: Construct a graph where nodes represent species and edges represent mass relationships derived from reactions (e.g., equality, "greater-than").
- Detect Contradictions: Traverse the graph to find cycles or paths that lead to a logical contradiction, such as a species being forced to have a mass greater than itself.
- Extract RIS and SIS: From the subgraph involved in the contradiction, extract the minimal set of reactions (RIS) and species (SIS) that are sufficient to produce the inconsistency.
Software Tools: The GAMES algorithm is also implemented within the SBMLLint package, which can be applied to curated models from the BioModels repository [3].
Table 2: Comparison of Two Primary Structural Error Isolation Protocols
| Aspect | Moiety Analysis | GAMES Algorithm |
|---|---|---|
| Primary Objective | Detect chemical group transfer errors. | Identify stoichiometric mass contradictions. |
| Level of Analysis | Chemical structure/functional groups. | Network topology and mass relationships. |
| Handles Implicit Molecules | Yes, moieties can be implicit. | No, relies on explicit stoichiometric relationships. |
| Core Output | List of reactions with moiety imbalance. | A minimal (RIS, SIS) pair explaining the inconsistency. |
| Ideal Use Case | Verifying transferase reactions, metabolic pathways. | Debugging large-scale network models pre-simulation. |
Visualizing Structural Errors and Isolation Sets
The following diagram, generated using Graphviz, illustrates how a stoichiometric inconsistency arises from a small set of reactions and how the GAMES algorithm isolates the relevant RIS and SIS.
Diagram 1: Isolation of a Stoichiometric Inconsistency. This graph visualizes a simplified inconsistency isolated by the GAMES algorithm. Reactions v537 and v601 enforce mass equality between species, placing c10, c154, and c160 in the same mass equivalence set (green). However, reaction v13 implies that the mass of c160 is greater than that of c10 or c154. Since all three species must have equal mass, a contradiction arises where c160 must be greater than itself. The RIS (red) and SIS (blue) neatly encapsulate the entire error.
Table 3: Key Research Reagent Solutions for Structural Error Analysis
| Tool / Resource | Function | Usage in Error Isolation |
|---|---|---|
| SBMLLint | Open-source linting tool for SBML models. | Implements both moiety analysis and the GAMES algorithm to detect and isolate structural errors [3]. |
| BioModels Repository | Public repository of curated, computational models of biological processes. | Source of real-world models (e.g., BIOMD0000000255) for testing and validating error isolation methods [3]. |
| Systems Biology Markup Language (SBML) | Open standard format for representing computational models in systems biology. | Provides a standardized, machine-readable format that enables the development and application of tools like SBMLLint [3]. |
| Stoichiometric Matrix | Mathematical representation of the reaction network. | The fundamental data structure analyzed by both linear programming methods and the GAMES algorithm to detect inconsistencies. |
| Moiety Annotation | Metadata defining chemical structures within species. | Essential input for performing moiety analysis; can be derived from chemical databases or manual curation. |
The isolation of structural errors through the precise definition of Reaction and Species Isolation Sets (RIS/SIS) represents a significant advancement in the construction of reliable biochemical models. By moving from generic error detection to targeted error explanation, methodologies like moiety analysis and the GAMES algorithm directly address the research question of how stoichiometric inconsistencies create critical gaps in reaction networks. These gaps are not merely inconveniences but fundamental logical flaws that undermine a model's validity. For researchers and drug development professionals, the adoption of these isolation techniques is paramount. It enables efficient debugging of complex models, enhances the credibility of models drawn from public repositories, and ultimately accelerates the development of accurate predictive models in systems biology.
Addressing Dead-End Metabolites and Orphan Reactions
Genome-scale metabolic reconstructions are biochemically, genetically, and genomically (BiGG) structured knowledge bases that formally represent the known metabolic activities of an organism [34]. These reconstructions are built from annotated genomic data and can be converted into constraint-based models for computational analysis. However, even the most complete models contain inconsistencies known as network gaps, which severely limit their predictive accuracy and biological relevance [34] [35]. These gaps manifest primarily as dead-end metabolites and orphan reactions, both of which disrupt metabolic connectivity and represent critical missing knowledge in our understanding of cellular biochemistry.
The presence of network gaps is fundamentally intertwined with the challenge of stoichiometric inconsistency, which occurs when the stoichiometry of reactions cannot satisfy mass conservation principles [19]. This creates biologically impossible scenarios where metabolites appear or disappear without balanced chemical equations. Within the context of broader research on how stoichiometric inconsistency creates network gaps, this technical guide provides comprehensive methodologies for identifying, analyzing, and resolving these critical model deficiencies to build more accurate and predictive metabolic networks.
Classifying and Detecting Network Gaps
Types of Missing Information in Metabolic Networks
Dead-End Metabolites: These metabolites have either producing or consuming reactions missing, creating termination points in metabolic pathways [34] [35]. They are further classified as:
- Root No-Production (RNP) metabolites: Only consumed by network reactions but never produced [35].
- Root No-Consumption (RNC) metabolites: Only produced by network reactions but never consumed [35].
- Downstream Non-Produced (DNP) metabolites: Become gaps as a consequence of upstream RNP metabolites [35].
- Upstream Non-Consumed (UNC) metabolites: Become gaps as a consequence of downstream RNC metabolites [35].
Orphan Reactions: These are biochemical reactions known to occur based on experimental evidence, but their catalytic genes remain unidentified [34]. Orphan reactions represent a significant challenge in metabolic reconstruction as they reflect limitations in both genomic annotation and biochemical characterization.
Blocked Reactions: Reactions that cannot carry steady-state flux under any physiological conditions due to connectivity issues within the network [35]. These reactions often occur downstream or upstream of dead-end metabolites and form isolated subnetworks within the broader metabolic network.
Detection Methodologies
Constraint-Based Analysis provides the mathematical foundation for identifying network gaps through stoichiometric matrix analysis [35]. The stoichiometric matrix (N) represents all metabolic reactions, with rows corresponding to metabolites and columns to reactions. Dead-end metabolites can be detected by scanning rows of the stoichiometric matrix for metabolites that appear only as reactants or only as products across all reactions [35].
The flux space (F) is defined using the steady-state mass balance equation and flux constraints:
where v represents flux distributions, and lower/upper bounds constrain reaction directions and capacities [35].
A reaction is classified as blocked if it cannot maintain a steady-state flux other than zero:
Propagation analysis identifies how the absence of flow through RNP or RNC metabolites affects downstream and upstream reactions, leading to DNP and UNC metabolites [35]. Advanced implementations such as the fastGapFill algorithm efficiently identify blocked reactions through a series of L1-norm regularized linear programs that approximate cardinality functions to identify compact flux-consistent models [19].
Computational Gap-Filling Strategies
Algorithmic Approaches
Table 1: Computational Gap-Filling Methods
| Method | Gap Type Addressed | Required Data | Key Features | References |
|---|---|---|---|---|
| fastGapFill | Dead-end metabolites | Universal reaction database (e.g., KEGG) | Computationally efficient; handles compartmentalized models; identifies stoichiometric inconsistencies | [19] |
| GapFill | Dead-end metabolites | Database of potential reactions (e.g., MetaCyc) | Identifies minimal reaction sets to enable flux through blocked reactions | [34] |
| SMILEY | Dead-end metabolites | Growth phenotype data, reaction database | Uses experimental phenotype data to constrain gap-filling solutions | [34] |
| GrowMatch | Dead-end metabolites | Gene essentiality data, reaction database | Integrates gene essentiality data with gap-filling | [34] |
| OptFill | Dead-end metabolites | Universal reaction database | Holistic approach; avoids thermodynamically infeasible cycles (TICs) | [6] |
| BNICE | Dead-end metabolites | Generalized enzyme reaction rules | Generates novel biochemical transformations using reaction rules | [34] |
| SEED | Orphan reactions | Annotated genome sequences from other organisms | Comparative genomics to assign genes to orphan reactions | [34] |
Implementation Workflows
The fastGapFill algorithm implements a sophisticated multi-step workflow for gap-filling [19]:
Preprocessing: A cellularly compartmentalized metabolic model (S) without blocked reactions (B) is expanded by a universal metabolic database (U), with a copy placed in each cellular compartment to generate SU.
Transport Reaction Addition: For each metabolite in non-cytosolic compartments, reversible intercompartmental transport reactions are added. For extracellular metabolites, exchange reactions are added (reaction set X).
Global Model Construction: The sum of reaction sets is added to SU to generate a global model, which is extended with solvable blocked reactions (Bs).
Core Set Identification: All reactions of S and Bs represent the core set for the subsequent consistency analysis.
Compact Subnetwork Computation: The algorithm computes a subnetwork containing all core reactions plus a minimal number of reactions from the universal database, ensuring all reactions in the resulting compact subnetwork are flux-consistent.
The OptFill method introduces an optimization-based multi-step approach that performs thermodynamically infeasible cycle (TIC)-avoiding whole-model gapfilling, addressing a critical limitation of earlier methods that could introduce energy-generating cyclic routes [6].
Figure 1: Workflow of Computational Gap-Filling Algorithms
Stoichiometric Consistency and Network Gaps
The Foundation of Stoichiometric Consistency
Stoichiometric inconsistency represents a fundamental challenge in metabolic modeling, occurring when reaction stoichiometries violate mass conservation principles [19]. For example, consider the reactions:
These reactions are stoichiometrically inconsistent, as no positive molecular mass can be assigned to A, B, and C such that mass is balanced on both sides of both reactions [19]. Such inconsistencies create biologically impossible scenarios and propagate gaps throughout the network.
Stoichiometric inconsistencies often arise from database errors, incomplete pathway knowledge, or incorrect assignment of reaction directions. The fastGapFill algorithm incorporates a stoichiometric consistency check using approximate cardinality maximization to compute a maximal set of metabolites involved in reactions that conserve mass [19]. This approach helps identify and exclude stoichiometrically inconsistent reactions during the gap-filling process, preventing the introduction of thermodynamically impossible solutions.
Impact on Network Connectivity
Stoichiometric inconsistencies directly create and exacerbate network gaps through several mechanisms:
Dead-End Propagation: Inconsistent stoichiometries can transform potentially connected metabolites into dead-ends by disrupting balanced production/consumption relationships.
Reaction Blocking: Even when all necessary enzymes are present, stoichiometric inconsistencies can prevent flux through entire pathway segments, creating effectively blocked reactions.
False Gap Identification: Apparent dead-end metabolites may result from stoichiometric miscalculations rather than genuine missing biochemistry.
The application of consistency checking during gap-filling, as implemented in fastGapFill, ensures that proposed gap-filling solutions maintain stoichiometric balance, addressing the root cause of many network gaps rather than merely treating their symptoms [19].
Experimental Validation and Integration
Integrating Experimental Data
Computational gap-filling predictions require experimental validation to confirm their biological relevance. Several methodologies integrate experimental data with gap-filling approaches:
Growth Phenotype Data: SMILEY utilizes growth phenotype data (e.g., from Biolog assays) to constrain gap-filling solutions, ensuring predicted reactions enable observed growth capabilities [34].
Gene Essentiality Data: GrowMatch incorporates gene essentiality data to identify gaps by comparing model predictions with experimental essentiality results, then proposing reactions that resolve these discrepancies [34].
Metabolic Flux Data: OMNI uses metabolic flux data combined with reaction databases to identify missing reactions necessary to explain observed flux distributions [34].
Validation Case Studies
Table 2: Experimental Validations of Gap-Filling Predictions
| Discovery/Application | Gap-Filling Method | Validation Methods | Organism |
|---|---|---|---|
| putP as propionate transporter | SMILEY | Gene knockout phenotype, RT-PCR | E. coli |
| idnT as 5-keto-D-gluconate transporter | SMILEY | Gene knockout phenotype, RT-PCR | E. coli |
| dctA, yeaU, yeaT as D-malate uptake genes | SMILEY | Gene knockout phenotypes, RT-PCR, enzyme assay | E. coli |
| Pyrimidine catabolism pathway | SEED | Gene knockout phenotypes, GC/MS | Multiple bacteria |
| 86 new reactions for metabolic model | GapFill and GrowMatch | Improved gene essentiality predictions | M. genitalium |
| Refinement of metabolic reconstruction | GapFill and GrowMatch | Improved growth phenotype predictions | Salmonella |
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Research Materials for Gap-Filling Studies
| Reagent/Resource | Function/Application | Example Sources |
|---|---|---|
| Universal Reaction Databases | Source of candidate reactions for gap-filling | KEGG, MetaCyc, BiGG |
| Gene Essentiality Data | Validation of model predictions and gap identification | Published mutant libraries |
| Growth Phenotype Arrays | High-throughput assessment of metabolic capabilities | Biolog plates |
| Metabolite Standards | Identification and quantification of metabolic intermediates | Commercial suppliers |
| Enzyme Assay Kits | Validation of predicted enzymatic activities | Commercial suppliers |
| Genome-Scale Model Testing Frameworks | Validation of model predictions against experimental data | COBRA Toolbox |
| Glycosidase-IN-2 | Glycosidase-IN-2, MF:C13H23NO5, MW:273.33 g/mol | Chemical Reagent |
Advanced Applications and Future Directions
Specialized Applications
Obligate Endosymbiont Models: The analysis of dead-end metabolites in minimized metabolic networks, such as those of obligate endosymbionts, requires specialized approaches. In these systems, metabolic redundancies with hosts lead to loss of enzymatic steps, creating obligate metabolic complementation [35]. These shared metabolic abilities manifest as interrupted pathways when reconstructing endosymbiont networks. The application of gap-filling methods to the Blattabacterium cuenoti model (iCG238) enabled detection of unconnected modules and curation to produce the improved iMP240 model [35].
Metabolomics Integration: Tools such as MetaboAnalyst support functional analysis of untargeted metabolomics data, enabling the identification of pathway-level activities that can inform gap-filling decisions [36]. The "MS Peaks to Pathways" module supports functional interpretation of high-resolution mass spectrometry data for over 120 species, facilitating the connection between experimental metabolomics and computational gap-filling [36].
Emerging Methodologies
Multi-Omics Integration: Next-generation gap-filling approaches increasingly incorporate transcriptomic, proteomic, and metabolomic data to constrain solution spaces and improve biological relevance. MetaboAnalyst provides joint pathway analysis capabilities that enable simultaneous analysis of gene and metabolite lists, supporting more comprehensive network gap identification [36].
Thermodynamic Constraining: Methods such as OptFill address the critical issue of thermodynamically infeasible cycles (TICs) that can emerge during automated gap-filling [6]. By incorporating thermodynamic constraints directly into the gap-filling optimization, these approaches produce more biologically plausible solutions that avoid energy-generating cyclic routes.
Figure 2: Relationship Between Stoichiometric Inconsistency and Network Gaps
Addressing dead-end metabolites and orphan reactions represents a critical challenge in metabolic network reconstruction and refinement. The integration of sophisticated computational algorithms such as fastGapFill and OptFill with experimental validation provides a powerful framework for identifying and resolving network gaps. Fundamental to this process is recognizing the role of stoichiometric inconsistency in creating and propagating these gaps through disruption of mass conservation principles.
As metabolic modeling continues to advance, the development of increasingly integrated approaches that combine multi-omics data, thermodynamic constraints, and automated curation will further enhance our ability to build biologically accurate metabolic networks. These improvements will directly support applications in biotechnology, biomedical research, and drug development by providing more predictive models of cellular metabolism.
Stoichiometric inconsistency, referring to imbalances in the elemental and reaction composition of biological systems, creates critical gaps in computational models of biological networks. These gaps manifest as missing reactions in metabolic networks, incorrect elemental ratios in organismal stoichiometry, and flawed predictions in synthetic biology applications. The core problem lies in the disconnect between theoretical computational models and experimentally viable biological states, which hampers drug development, metabolic engineering, and functional genomics research. As research increasingly relies on in silico predictions to guide laboratory experimentation, developing robust optimization techniques to prioritize biologically relevant solutions has become paramount for researchers and drug development professionals.
The fundamental challenge arises from the complex nature of biological systems where multiple competing models often satisfy limited experimental data equally well [37]. For instance, in metabolic network analysis, the stoichiometry matrix invariably contains redundancies that reflect dependencies within the network [38]. Similarly, in materials science, most candidate materials identified computationally prove impractical to synthesize, creating a significant gap between prediction and reality [15]. This whitepaper examines computational optimization techniques that bridge this divide by prioritizing solutions consistent with biological constraints, experimental data, and synthetic feasibility.
Technical Approaches to Optimization
Machine Learning for Synthesizability Prediction
Machine learning approaches have revolutionized the prediction of biologically feasible solutions by learning hidden patterns from existing biological data. The CHESHIRE (CHEbyshev Spectral HyperlInk pREdictor) method exemplifies this approach for gap-filling in Genome-scale Metabolic Models (GEMs) by predicting missing reactions purely from metabolic network topology [16]. This deep learning-based method employs a sophisticated architecture with four major steps: feature initialization, feature refinement using Chebyshev spectral graph convolutional network (CSGCN), pooling to integrate metabolite-level features, and scoring to produce probabilistic confidence scores for reaction existence [16].
Table 1: Performance Comparison of Topology-Based Gap-Filling Methods
| Method | AUROC | Key Principle | Data Requirements |
|---|---|---|---|
| CHESHIRE | 0.89 | Hypergraph learning with spectral graph convolution | Network topology only |
| NHP | 0.85 | Neural hyperlink prediction with graph approximation | Network topology only |
| C3MM | 0.82 | Clique closure-based matrix minimization | Network topology only |
| Node2Vec-mean | 0.79 | Random walk embedding with mean pooling | Network topology only |
In materials stoichiometry, semi-supervised learning using positive-unlabeled learning achieves a true positive rate of 83.4% and precision of 83.6% for predicting synthesizable inorganic materials [15]. This approach learns hidden features of synthesizable compositions and allows construction of continuous synthesizability phase maps aligned with available synthetic data, successfully guiding experimental exploration of quaternary oxide compositional space to discover new phases like Cuâ‚„FeV₃Oâ‚₃ [15].
Constraint Satisfaction and Multi-Objective Optimization
Constraint Satisfaction Problems (CSP) provide a declarative and efficient framework for describing combinatorial problems in biological networks that must satisfy broad constraint sets [37]. The Biological Model Checker (Bio-ModelChecker) implements this approach using bounded constraint satisfaction to identify parameter sets for regulatory networks that comply with experimental observations [37]. This framework formulates model identification as a multi-objective optimization problem directed at maximizing structural parsimony by mitigating excessive control action selectivity while favoring increased state transition efficiency and robustness of the network's dynamic response [37].
Table 2: Optimization Criteria for Biological Model Ranking
| Criterion | Biological Interpretation | Mathematical Formulation |
|---|---|---|
| Selectivity | Mitigation of excessive control action specificity | Minimization of redundant network edges |
| Efficiency | Minimum transitions to reproduce measurement data | State transition path length optimization |
| Robustness | Stability of dynamic response to perturbations | Resistance to state transition failure |
| Parsimony | Minimal complexity consistent with data | Akaike Information Criterion (AIC) principles |
The power of CSP approaches lies in their ability to handle sparse, irregularly collected, and incompletely surveyed samples – common challenges in experimental biology [37]. By incorporating both synchronous and asynchronous discrete updating schemes, these methods can efficiently check whether a regulatory model reproduces time-sampled measurements while accommodating the exponential complexity of biological networks [37].
Stoichiometric Matrix Factorization and Network Reduction
Stoichiometric redundancies in metabolic networks can be exploited for computational efficiency through matrix factorization approaches [38]. The observation that the steady-state balance condition depends only on the form of redundancies within the stoichiometry matrix N enables significant simplification of steady-state analysis. The complete stoichiometric reduction approach factors the system dynamics as:
Where L is the row link matrix, NRC is the stoichiometric core, and P is the column link matrix [38]. This factorization allows for targeted elimination of specific reactions to generate simplified descriptions of flux profiles within input-output network modules, significantly reducing computational effort required for elementary flux mode analysis [38].
Experimental Protocols and Methodologies
Genome-Scale Metabolic Model Reconstruction and Validation
The reconstruction of genome-scale metabolic models (GEMs) follows a standardized protocol for determining metabolic capabilities of biological systems [39]. For Bartonella quintana str. Toulouse, the protocol begins with genome annotation using RAST (Rapid Annotation using Subsystem Technology) and ModelSEED to obtain a draft metabolic network [39]. This is followed by manual refinement using multiple databases including KEGG, BioCyc, and BRENDA to ensure accuracy in gap-filling. Reactions forming unconnected modules are removed to avoid inconsistencies, and the model quality is assessed using MEMOTE [39].
Flux Balance Analysis (FBA) is then performed to determine optimal metabolic flux distributions, maximizing the biomass reaction as an objective function under steady-state assumption [39]. For organisms without documented biomass composition, comparison with well-characterized models like E. coli iJO1366 provides a reference for adjustment. Experimental validation includes testing modified culture media based on model predictions (e.g., 2-oxoglutarate supplementation) and proteomic analyses to identify metabolic adaptations [39]. This protocol successfully identified key metabolic requirements in B. quintana, demonstrating the utility of GEMs for optimizing growth conditions of fastidious organisms.
Network Alignment and Consistency Checking
Network alignment methodology requires careful preprocessing to ensure biologically meaningful comparisons [40]. The protocol begins with robust identifier mapping and normalization using resources like UniProt, HGNC, or Ensembl to address gene/protein synonym challenges. Gene names are normalized across datasets using tools such as UniProt ID mapping, NCBI Gene, or MyGene.info API, with adoption of HGNC-approved gene symbols for human datasets and equivalent authoritative sources for other species [40].
The choice of network representation format critically impacts alignment efficiency and accuracy [40]. For protein-protein interaction networks, adjacency lists are preferred for memory efficiency and scalable traversal of typically large, sparse networks. For gene regulatory networks with denser interactions, adjacency matrices better support matrix-based operations and compact representation of pairwise relationships [40]. Metabolic networks, often directed and weighted, benefit from edge lists that offer flexible parsing and preserve path directionality. Following format selection, alignment algorithms optimize node and edge similarity, functional annotations, or topological features to generate alignments maximizing biological relevance [40].
Stoichiometric Data Compilation and Quality Control
The StoichLife global dataset compilation protocol provides a framework for ensuring data quality in stoichiometric studies [41]. The protocol involves developing a standardized template structure containing elemental content and ratios (%C, %N, %P, C:N, C:P, and N:P) alongside body size measurements, sampling locality, and taxonomic affiliation [41]. Data undergoes rigorous validation and quality assurance procedures with three distinct data types processed separately: quantitative, taxonomic, and spatial.
Quantitative data verification ensures elemental content values represent percentage of each element in dry body mass, with elemental ratios checked for accurate reflection of both mass and molar ratios [41]. Taxonomic data validation involves automated and manual inspection to correct spelling errors, complete missing information, address ambiguously identified morphospecies, and ensure accuracy of accepted names using GBIF, ITIS, and Catalogue of Life databases. Spatial data validation includes visual inspection by plotting coordinates onto global maps to identify and correct errors such as marine data mistakenly recorded as inland [41].
Table 3: Key Research Reagent Solutions for Stoichiometric Network Analysis
| Resource | Type | Function | Application Context |
|---|---|---|---|
| COBRApy | Python module | Metabolic modeling and analysis | GEM reconstruction and FBA [39] |
| ModelSEED | Database platform | Draft metabolic network generation | Automated metabolic reconstruction [39] |
| RAST | Annotation service | Genome annotation | Initial metabolic network drafting [39] |
| BiGG Models | Knowledgebase | Curated metabolic networks | GEM validation and benchmarking [16] |
| CHEESHIRE | Algorithm | Hyperlink prediction | Topology-based gap-filling in GEMs [16] |
| Bio-ModelChecker | Software tool | Constraint satisfaction checking | Regulatory network parameterization [37] |
| StoichLife | Database | Organismal stoichiometry data | Ecological stoichiometry studies [41] |
| MEMOTE | Test suite | GEM quality assessment | Metabolic model validation [39] |
Optimization techniques that prioritize biologically relevant solutions represent a paradigm shift in computational biology, bridging the critical gap between theoretical predictions and experimental reality. By leveraging machine learning for synthesizability prediction, constraint satisfaction for multi-objective optimization, and stoichiometric matrix factorization for network reduction, researchers can significantly enhance the biological relevance of their computational models. The continued refinement of these approaches, supported by standardized experimental protocols and comprehensive reagent toolkits, promises to accelerate drug development, metabolic engineering, and synthetic biology applications by ensuring computational efforts yield experimentally viable solutions consistent with biological constraints.
The reconstruction of predictive, genome-scale metabolic models (GEMs) is a cornerstone of systems biology, with applications ranging from metabolic engineering to drug target identification [42] [43]. The foundational premise of these models is the principle of mass conservation, mathematically represented by the stoichiometric matrix S, where Sv = 0 defines the system at steady state. The transition from modeling individual microbes to complex human-scale systems, including diverse microbial communities like the gut microbiome, magnifies a fundamental problem: stoichiometric inconsistency [44] [3].
These inconsistencies—violations of mass balance and energy conservation—create critical network gaps that cripple the predictive power of in silico models. In a single microbial model, an error might affect a single pathway; in a community-scale human model, that same error propagates, creating cascading failures that render simulations of host-microbiome-diet interactions unreliable [44] [43]. This whitepaper examines how stoichiometric inconsistencies arise, their systemic impact on model scaling, and the methodologies for their identification and resolution, framed within the broader research on network gaps.
Stoichiometric Inconsistency: A Primary Source of Network Gaps
Defining Structural and Stoichiometric Errors
In reaction networks, a stoichiometric inconsistency is a structural error that implies one or more chemical species must have a mass of zero to satisfy the network's constraints, representing a logical impossibility [3]. These errors manifest in two primary forms:
- Mass and Atomic Imbalance: The most straightforward error occurs when the counts of individual atoms (e.g., C, N, O, P) in a reaction's reactants do not equal the counts in its products. Atomic Mass Analysis (AMA) is the standard method for detecting these errors [3].
- Moiety Imbalance: A more subtle but equally critical error involves the imbalance of conserved chemical groups or moieties (e.g., inorganic phosphate, adenosine) between reactants and products. Unlike atoms, the atomic composition of a moiety can vary slightly between molecular contexts. For example, in the reaction ATP → ADP + Pi, the reaction is moiety-balanced (one adenosine, three phosphates on each side) but not mass-balanced unless water is included [3].
The Mechanism of Gap Creation
Stoichiometric inconsistencies directly create network gaps by breaking the connectivity of the metabolic network. A reaction with a mass balance error cannot carry flux without violating physical laws, effectively becoming non-functional. The algorithms used for Flux Balance Analysis (FBA) and related methods depend on a continuous flow of mass through the network. Inconsistencies block this flow, leading to "dead-end" metabolites and "blocked" reactions that are incorrectly predicted to be unable to carry flux under any condition, even if the necessary enzymes are present [3] [6].
The following diagram illustrates the logical relationship between different types of stoichiometric errors and the network gaps they create.
Diagram 1: The causal pathway from stoichiometric errors to model failure.
The Scaling Problem: From Microbial to Community and Human Models
The challenges of managing stoichiometric inconsistencies intensify dramatically as model complexity increases.
Microbial Models: A Contained Challenge
In single-organism models, such as those for Corynebacterium glutamicum or Escherichia coli, the network is self-contained. Gap-filling can often be performed by referencing a well-defined set of biochemical transformations from databases like KEGG or MetaCyT [42] [6]. Tools like OptFill can perform holistic, infeasible cycle-free gapfilling for these systems, adding missing reactions to restore connectivity while avoiding the creation of thermodynamically infeasible loops (TICs) that are another form of structural error [6].
Community and Human Models: A Compounded Problem
Scaling to human gut microbiome models or human metabolic networks introduces several layers of complexity that compound the issue of stoichiometric inconsistency [44] [43].
- Multi-Compartmentalization: Human models require multiple cellular compartments (cytosol, mitochondria, nucleus, etc.), each with its own metabolite pools. Inconsistencies can arise from incorrect transport reactions between these compartments.
- Cross-Species Metabolite Exchange: In community models like the gut microbiome, the metabolic outputs of one organism become the inputs for another. A gap in one species' model can propagate, causing cascading failures in the simulated community [44] [43].
- Multi-Objective Optimization: Simple steady-state FBA with a single objective (e.g., biomass maximization) may be insufficient. Community dynamics may require considering multiple, competing objectives, which demands higher computational cost and increases the potential for solutions that are mathematically feasible but biologically inconsistent [44].
- Integration with Host Metabolism: The ultimate challenge is integrating microbial community models with the human host. Inconsistencies in either sub-model can invalidate predictions of host-microbiome interactions, which is critical for applications like Live Biotherapeutic Products (LBP) development [43].
Table 1: Comparative Challenges in Model Scaling
| Aspect | Microbial Model (e.g., E. coli) | Community/Human Model (e.g., Gut Microbiome) |
|---|---|---|
| System Boundary | Single cell, defined compartments | Multiple species, multiple host compartments |
| Mass Balance Scope | Self-contained biochemistry | Cross-species metabolite exchange |
| Primary Gap-Filling | Internal pathway completion | Inter-species and host-pathway integration |
| Computational Demand | Relatively low | High; requires sophisticated algorithms [44] |
| Key Tool Example | OptFill [6] | AGORA2 framework [43] |
Experimental and Computational Protocols for Validation
Robust model validation requires a combination of computational linting and experimental verification.
Computational Error Isolation and Gap-Filling
Protocol 1: Structural Error Linting with SBMLLint
- Objective: Identify and isolate mass and moiety balance errors in a model encoded in SBML.
- Methodology:
- Input: A metabolic model in SBML format.
- Moiety Analysis: Run the model through a linter like SBMLLint, which uses algorithms to decompose species into moieties and checks for their conservation in reactions, independent of atomic formulas [3].
- Error Isolation: Apply the Graphical Analysis of Mass Equivalence Sets (GAMES) algorithm. GAMES constructs explanations that pinpoint a small set of reactions (Reaction Isolation Set - RIS) and species (Species Isolation Set - SIS) responsible for stoichiometric inconsistencies, simplifying error remediation [3].
- Output: A report listing unbalanced reactions and a minimal set of elements causing structural errors.
Protocol 2: TIC-Avoiding Gap-Filling with OptFill
- Objective: Automatically fill network gaps in a genome-scale model without introducing thermodynamically infeasible cycles.
- Methodology:
- Input: An incomplete stoichiometric model and a biochemical reaction database.
- Multi-Step Optimization: OptFill uses an optimization-based method to select a set of reactions from the database to add to the model. This method is "holistic," considering the entire network rather than one metabolite at a time [6].
- TIC Prevention: The algorithm is explicitly designed to ensure the gap-filled solution does not introduce TICs, a common problem with other gap-filling tools that necessitates manual curation [6].
- Output: A complete, gap-free, and thermodynamically consistent metabolic model.
Experimental Validation of Model Predictions
Protocol 3: Metabolomic Validation of Flux Predictions
- Objective: Validate in silico flux predictions using experimental metabolomics to ensure model consistency with biological reality.
- Methodology:
- In Silico Prediction: Use FBA or FVA on the curated model to predict extracellular metabolite secretion/uptake rates or intracellular flux distributions under defined growth conditions [42] [43].
- Cultivation: Grow the organism(s) in a controlled bioreactor under the same conditions used in the simulation.
- Metabolite Profiling: Use mass spectrometry-based platforms like MetaboAnalyst to quantify extracellular metabolite concentrations in the spent media and/or perform intracellular metabolomics [36]. LC-MS Spectral Processing and Statistical Analysis modules within MetaboAnalyst can be used for this purpose.
- Data Integration: Calculate experimental exchange fluxes from concentration changes and compare them to model predictions. Significant discrepancies indicate potential remaining network gaps or incorrect constraints.
The workflow below integrates these computational and experimental methods into a cohesive model refinement cycle.
Diagram 2: An integrated workflow for model curation and validation.
Table 2: Key Research Reagent Solutions for Metabolic Modeling and Validation
| Tool/Resource | Type | Primary Function | Relevance to Stoichiometric Consistency |
|---|---|---|---|
| SBMLLint [3] | Software Library | Static error checker ("linter") for reaction networks | Detects mass & moiety imbalances; isolates errors via GAMES algorithm. |
| AGORA2 [43] | Model Database | Repository of 7,302 curated, strain-level GEMs for gut microbes | Provides a reference for stoichiometrically consistent community modeling. |
| OptFill [6] | Gap-filling Algorithm | Optimization-based tool for model completion | Adds missing reactions to close network gaps without creating TICs. |
| Metano/MMTB [42] | Modeling Toolbox | Software for flux analysis (FBA, FVA, MOMA) | Enables metabolite-centric view (via MFM) to analyze flux distributions and identify inconsistencies. |
| MetaboAnalyst [36] | Web-based Platform | Comprehensive metabolomics data analysis | Validates model predictions by comparing in-silico fluxes with experimental metabolomic data. |
| COBRA Toolbox [3] [42] | Modeling Toolkit | A standard platform for constraint-based reconstruction and analysis | Widely used for FBA, FVA, and includes mass balance checking capabilities. |
The path from microbial to human metabolic modeling is fraught with challenges rooted in the fundamental principle of mass conservation. Stoichiometric inconsistency is not merely a technical nuisance; it is a primary generator of network gaps that undermine model scalability and predictive accuracy. Addressing this requires a rigorous, multi-faceted approach: employing sophisticated "linter" tools for error detection and isolation, utilizing next-generation gap-filling algorithms that preempt thermodynamic errors, and adhering to a cycle of computational prediction and experimental validation. As the field moves toward personalized, community-scale models for therapeutic applications [44] [43], the development of robust, automated, and validated frameworks for ensuring stoichiometric consistency will be the cornerstone of reliable, translational systems biology research.
Benchmarking Success: Validating and Comparing Gap-Filling Solutions
In the field of systems biology, genome-scale metabolic models (GEMs) serve as powerful mathematical representations of cellular metabolism, predicting metabolic fluxes in living organisms through gene-reaction-metabolite connectivity [16]. The accuracy of these reaction-based models is paramount as they progressively advance various disciplines in biomedical sciences, including metabolic engineering, microbial ecology, and drug discovery [16] [3]. However, the growing complexity of reaction-based models, some containing thousands of reactions, necessitates rigorous error-checking methodologies [3]. Internal validation represents a critical approach for verifying model correctness by testing a method's ability to recover artificially introduced gaps—deliberately removed reactions from metabolic networks—before experimental data becomes available [16].
This validation approach operates within a broader research context investigating how stoichiometric inconsistencies create network gaps. Stoichiometric inconsistencies represent fundamental structural errors in reaction networks that imply one or more chemical species have a mass of zero, creating knowledge gaps that compromise model predictions [3]. For example, in one BioModels entry with 827 reactions, analysis revealed a contradiction where reaction relationships implied a chemical species must have a mass larger than itself—a clear logical impossibility stemming from structural errors [3]. Internal validation through artificial gap recovery provides a methodological framework for testing and refining computational tools designed to address these fundamental model integrity issues.
Theoretical Foundation: Stoichiometric Inconsistency as a Source of Network Gaps
The Nature of Structural Errors in Metabolic Networks
Stoichiometric inconsistencies represent a serious category of structural errors in reaction-based models. These inconsistencies arise when the stoichiometric relationships between reactions imply that one or more chemical species must have a mass of zero, creating logical impossibilities that propagate through the network [3]. Such errors typically originate from incorrect reaction specifications, where the transformation of reactants to products violates mass conservation principles or creates contradictory mass relationships between species.
A specific manifestation of structural errors occurs in the form of moiety imbalances, where chemical structures (moieties) become unbalanced between reactants and products [3]. Unlike atomic mass analysis, which compares atom counts in reactants and products, moiety analysis operates at a higher level of chemical abstraction, examining the balance of functional groups that may have slightly varying atomic compositions. For example, in ATP hydrolysis (ATP → ADP + Pi), the reaction is moiety balanced (one adenosine and three phosphates on both sides) but not mass balanced due to differences in atomic formulas of the inorganic phosphates [3]. These nuanced structural errors frequently create gaps that disrupt metabolic network functionality.
The Gap-Filling Challenge in Metabolic Models
The presence of network gaps, whether arising from stoichiometric inconsistencies or incomplete knowledge, presents significant challenges for metabolic modeling:
- Knowledge Gaps: Due to imperfect knowledge of metabolic processes, even highly curated GEMs contain missing reactions that must be identified and filled [16]
- Phenotypic Prediction Errors: Gaps disrupt accurate prediction of metabolic phenotypes, such as fermentation products and amino acid secretion [16]
- Cascade Failures: In highly coupled networks, a single gap can trigger cascading failures across interconnected systems [45] [46]
The process of "gap-filling" represents a crucial step in metabolic model refinement, aiming to identify and incorporate missing reactions to restore network functionality and improve predictive accuracy [16].
Methodological Framework: Internal Validation for Gap Recovery
Internal validation for testing gap-filling methods follows a systematic approach centered on artificially introducing gaps into known metabolic networks and evaluating recovery performance. The fundamental principle involves treating the metabolic network as a hypergraph where each hyperlink represents a metabolic reaction connecting participating reactant and product metabolites [16]. This representation naturally captures the multi-molecular nature of biochemical reactions.
The validation procedure follows these essential steps:
- Network Selection: Curated, high-quality metabolic models (e.g., from BiGG or BioModels repositories) serve as benchmark networks [16] [3]
- Artificial Gap Creation: Existing reactions are systematically removed from the network
- Recovery Attempt: Computational methods attempt to identify and restore the missing reactions
- Performance Quantification: Predictive accuracy is measured by comparing recovered reactions against the known, artificially removed reactions
This approach enables rigorous testing without requiring experimental phenotypic data, making it particularly valuable for non-model organisms where such data is unavailable [16].
Experimental Design: Implementing Artificial Gap Recovery
The internal validation process follows a detailed experimental workflow to ensure comprehensive assessment of gap-filling methodologies:
Diagram 1: Experimental workflow for internal validation with artificial gaps.
The process employs negative sampling to create realistic negative examples for model training and evaluation. For each positive reaction (existing in the metabolic network), a corresponding negative reaction is generated by replacing approximately half of the metabolites with randomly selected metabolites from a universal metabolite pool, maintaining a 1:1 positive-to-negative ratio [16]. This approach ensures the model learns to distinguish legitimate reactions from implausible ones.
Advanced Computational Methods for Gap-Filling
Hypergraph Learning Approaches
State-of-the-art methods for gap-filling leverage hypergraph learning frameworks to predict missing reactions:
- CHESHIRE (CHEbyshev Spectral HyperlInk pREdictor): A deep learning-based method that uses hypergraph topology to predict missing reactions through feature initialization, refinement via Chebyshev spectral graph convolutional network (CSGCN), pooling, and scoring [16]
- NHP (Neural Hyperlink Predictor): A neural network-based approach that approximates hypergraphs using graphs, potentially losing higher-order information [16]
- C3MM (Clique Closure-based Coordinated Matrix Minimization): An integrated training-prediction method with limited scalability for large reaction pools [16]
These machine learning methods frame the prediction of missing reactions in a GEM as a task of predicting hyperlinks on a hypergraph, leveraging the natural representation of metabolic networks where each reaction connects multiple metabolites [16].
Moiety Analysis for Structural Error Detection
Beyond complete missing reactions, moiety analysis addresses structural errors by detecting imbalances of chemical structures between reactants and products [3]. This approach uses the same algorithmic framework as atomic mass analysis but operates in units of moieties rather than individual atoms, capturing higher-level chemical structure preservation that may be missed by atomic-level analysis.
Performance Metrics and Comparative Analysis
Quantitative Evaluation Metrics
Internal validation relies on established classification performance metrics to quantitatively assess gap-filling methods:
- AUROC (Area Under the Receiver Operating Characteristic curve): Measures the ability to distinguish between true positive reactions and negative reactions across all classification thresholds [16]
- Precision-Recall Metrics: Evaluate the trade-off between correctly identified missing reactions (true positives) and incorrect predictions (false positives)
- Recovery Rate: The proportion of artificially removed reactions that are correctly identified and restored
These metrics provide complementary insights into method performance, with AUROC particularly valuable for evaluating the ranking capability of predictive algorithms.
Comparative Performance of Gap-Filling Methods
Table 1: Performance comparison of topology-based gap-filling methods on BiGG models
| Method | Architecture | Key Features | AUROC | Limitations |
|---|---|---|---|---|
| CHESHIRE | Deep learning | Chebyshev spectral graph convolutional network, hypergraph topology | Highest | Complex architecture requiring substantial computational resources |
| NHP (Neural Hyperlink Predictor) | Neural network | Graph approximation of hypergraphs | Intermediate | Loss of higher-order information from hypergraph simplification |
| C3MM | Matrix minimization | Integrated training-prediction, clique closure | Lower | Limited scalability for large reaction pools |
| Node2Vec-mean | Graph embedding | Random walk-based node features, mean pooling | Baseline | Simple architecture without feature refinement |
Table 2: Validation outcomes for CHESHIRE across different model types
| Model Type | Number Tested | Validation Type | Key Outcome | Application Context |
|---|---|---|---|---|
| BiGG Models | 108 | Internal validation (artificial gaps) | Outperformed other topology-based methods | High-quality curated metabolic networks |
| AGORA Models | 818 | Internal validation (artificial gaps) | Superior recovery performance | Microbial metabolic models |
| Draft GEMs | 49 | External validation (phenotypic prediction) | Improved predictions of fermentation products & amino acid secretion | Automatically reconstructed models |
Experimental Protocols for Internal Validation
Standard Protocol for Artificial Gap Recovery
A comprehensive internal validation experiment follows this detailed protocol:
Model Selection and Preparation
- Select 108 high-quality BiGG models or 818 AGORA models as benchmark datasets [16]
- Represent the metabolic network as a hypergraph where reactions are hyperlinks connecting metabolite nodes
Training-Testing Split
- Split metabolic reactions into training (60%) and testing (40%) sets over 10 Monte Carlo runs to ensure statistical robustness [16]
- This cross-validation approach accounts for variability in reaction selection
Artificial Gap Introduction
- Remove reactions from the testing set to simulate network gaps
- Generate negative reactions at 1:1 ratio to positive reactions by replacing half (rounded if needed) of metabolites in positive reactions with randomly selected metabolites from a universal metabolite pool [16]
Model Training and Prediction
- Train models on the training set combined with generated negative reactions
- Test model performance on the testing set mixed with either derived negative reactions or real reactions from a universal database [16]
Performance Evaluation
- Calculate AUROC values and other classification metrics
- Compare performance against state-of-the-art methods (NHP, C3MM, Node2Vec-mean)
Validation with Phenotypic Predictions
Beyond internal validation with artificial gaps, external validation assesses the biological relevance of gap-filling through phenotypic prediction accuracy:
- Test the ability of curated models to predict fermentation products and amino acid secretion [16]
- Compare phenotypic predictions before and after gap-filling to quantify improvement
- Use draft GEMs from automated reconstruction pipelines (CarveMe, ModelSEED) as test cases [16]
Research Reagent Solutions for Metabolic Network Analysis
Table 3: Essential research tools and resources for metabolic network gap analysis
| Resource Type | Specific Tools/Databases | Function | Application in Gap-Filling |
|---|---|---|---|
| Model Repositories | BiGG Models, BioModels, AGORA | Provide curated metabolic models for benchmarking | Source of high-quality models for training and testing gap-filling methods |
| Analysis Toolkits | COBRA Toolbox, MEMOTE | Enable metabolic network analysis and validation | Implement mass balance checking and network validation |
| Structural Analysis | SBMLLint, Moisty Analysis Tools | Detect stoichiometric inconsistencies and moiety imbalances | Identify structural errors that create network gaps [3] |
| Computational Frameworks | CHESHIRE, NHP, C3MM | Predict missing reactions in metabolic networks | Direct implementation of gap-filling algorithms [16] |
| Chemical Databases | Universal metabolite databases | Source of candidate metabolites for negative sampling | Generate plausible negative reactions for model training [16] |
Implications for Drug Discovery and Development
Robust internal validation methodologies for gap recovery in metabolic networks have significant implications for pharmaceutical research and development:
- Target Identification: Complete metabolic networks enable more accurate identification of essential metabolic pathways in pathogens, revealing novel drug targets [16]
- Toxicity Prediction: Comprehensive models improve prediction of drug metabolite toxicity by ensuring complete representation of metabolic transformations [16]
- Microbiome Therapeutics: Enhanced gap-filling facilitates modeling of microbial communities relevant to human health, supporting microbiome-based therapeutic development [47]
- Network Pharmacology: Complete metabolic networks support systems-level understanding of drug action across interconnected metabolic pathways
The application of advanced gap-filling methods like CHESHIRE has demonstrated tangible improvements in predicting metabolic phenotypes, directly impacting drug discovery pipelines that rely on accurate metabolic modeling [16].
Internal validation through artificial gap recovery represents a rigorous methodology for advancing metabolic network completeness and correctness. By systematically testing the ability of computational methods to recover artificially removed reactions, researchers can refine gap-filling algorithms before experimental validation. The integration of hypergraph learning with moiety balance analysis addresses both complete reaction gaps and subtle structural errors stemming from stoichiometric inconsistencies.
As metabolic modeling continues to expand into non-model organisms and complex microbial communities, robust internal validation frameworks will become increasingly critical for ensuring model reliability in biomedical applications. The continuing development of methods like CHESHIRE that leverage topological features without requiring phenotypic data will accelerate the creation of high-quality metabolic models for drug discovery and therapeutic development.
Stoichiometric inconsistency—a disconnect between the theoretical capabilities of a metabolic network and its actual functional requirements—is a primary source of network gaps in genome-scale metabolic models (GEMs). These gaps manifest as missing reactions that prevent models from simulating observed metabolic phenotypes. For over a decade, optimization-based methods like fastGapFill have been the standard for gap-filling. Recently, topology-based machine learning (ML) methods such as CHESHIRE (CHEbyshev Spectral HyperlInk pREdictor) and NHP (Neural Hyperlink Predictor) have emerged as powerful alternatives. This whitepaper provides a comparative analysis of these paradigms, detailing their core principles, experimental validations, and performance. We demonstrate that while fastGapFill relies on a universal reaction database to force functional consistency, ML methods like CHESHIRE learn the underlying topological "blueprint" of metabolic networks to predict missing links, offering superior accuracy without dependency on experimental phenotypic data.
A Genome-scale Metabolic Model (GEM) is a mathematical representation of an organism's metabolism, encapsulating the relationships between genes, reactions, and metabolites through two key matrices: the stoichiometric matrix (associating metabolites with reactions) and the reaction-gene matrix (associating reactions with their corresponding enzymes) [16]. GEMs are powerful tools for predicting metabolic fluxes and understanding cellular physiology.
However, due to imperfect biological knowledge and incomplete genomic annotations, even highly curated GEMs contain knowledge gaps, most commonly missing reactions [16]. These gaps create stoichiometric inconsistencies—violations of mass-balance and energy conservation principles that render the network unable to produce or consume certain metabolites (dead-end metabolites) or to simulate experimentally observed growth and secretion profiles [48] [38]. From a computational perspective, these inconsistencies mean the stoichiometric matrix lacks the necessary columns (reactions) to represent a continuous flow of mass and energy, leading to incorrect flux balance analysis (FBA) predictions [38].
The fundamental challenge in gap-filling is to identify the minimal set of reactions whose addition resolves these stoichiometric inconsistencies and restores network functionality. The two paradigms discussed herein—fastGapFill and machine learning approaches—address this challenge through fundamentally different philosophies and methodologies.
Core Methodologies and Algorithms
fastGapFill: An Optimization-Based Approach
fastGapFill is a classic optimization-based gap-filling method that relies on the availability of a comprehensive universal reaction database (e.g., KEGG, MetaCyc) [16]. Its objective is to find the most parsimonious set of reactions from this database that, when added to an incomplete draft GEM, resolve dead-end metabolites and enable the simulation of a target metabolic function, such as biomass production [16] [48].
The core algorithm of fastGapFill can be summarized as a mixed-integer linear programming (MILP) problem:
- Problem Formulation: The draft network is represented by its stoichiometric matrix ( S{draft} ). A universal database of candidate reactions forms another stoichiometric matrix ( S{db} ). The combined network is ( S{combined} = [S{draft} | S_{db}] ).
- Objective Function: The goal is to minimize the number of added reactions from the database. This is achieved by introducing binary variables ( yi ) for each candidate reaction ( i ) in ( S{db} ), indicating whether the reaction is included.
- Constraints:
- Mass Balance: ( S{combined} \cdot v = 0 ), where ( v ) is the flux vector. This ensures steady-state operation.
- Reaction Capacity: ( lb \leq v \leq ub ), defining lower and upper flux bounds for each reaction.
- Target Function: A specific metabolic output is defined, e.g., ( v{biomass} \geq v{target} ), forcing the network to achieve a minimal level of a key function.
- Coupling Constraints: ( vi \leq ub \cdot yi ), ensuring that a flux through a candidate reaction is only possible if it is selected (( yi = 1 )).
- Solution: The MILP solver identifies the set of reactions ( {i | yi = 1} ) that minimizes the objective function ( \sum yi ) while satisfying all constraints.
Figure 1: The fastGapFill optimization workflow. The method integrates a draft model with a universal database and uses a Mixed-Integer Linear Programming (MILP) approach to find the minimal set of reactions that restore network functionality.
Machine Learning Approaches: CHESHIRE and NHP
ML methods like CHESHIRE and NHP frame gap-filling as a hyperlink prediction problem on a hypergraph, requiring only the topological structure of the metabolic network for training [16] [27].
- Metabolic Network as a Hypergraph: In this representation, each metabolite is a node, and each reaction is a hyperlink (edge) connecting all its reactant and product metabolites [16]. This naturally captures the higher-order interactions inherent to biochemical reactions, which cannot be fully represented in simple graphs where edges connect only two nodes.
CHESHIRE (CHEbyshev Spectral HyperlInk pREdictor) is a deep learning method designed to overcome the limitations of earlier ML approaches. Its architecture consists of four major steps [16]:
- Feature Initialization: An encoder-based one-layer neural network generates an initial feature vector for each metabolite from the hypergraph's incidence matrix (a boolean matrix indicating metabolite participation in reactions).
- Feature Refinement: A Chebyshev Spectral Graph Convolutional Network (CSGCN) operates on a "decomposed graph" (where each reaction is represented as a fully connected subgraph of its metabolites). The CSGCN refines each metabolite's feature vector by incorporating features from other metabolites in the same reaction, capturing complex metabolite-metabolite interactions.
- Pooling: For each reaction, the feature vectors of its participating metabolites are integrated into a single reaction-level feature vector. CHESHIRE combines two pooling functions: a max-min function and a Frobenius norm-based function, to provide complementary information.
- Scoring: The reaction-level feature vector is fed into a one-layer neural network to produce a probabilistic score indicating the confidence of the reaction's existence.
NHP (Neural Hyperlink Predictor) shares a similar architecture but approximates hypergraphs using simple graphs for node feature generation, which can lead to a loss of higher-order information. It also uses a simpler pooling strategy [16].
Figure 2: The CHESHIRE deep learning workflow. The model learns from the network topology to predict missing reactions, without requiring phenotypic data or a universal reaction database during training.
Paradigm Comparison: Key Philosophical Differences
Table 1: Core philosophical and operational differences between fastGapFill and ML approaches like CHESHIRE.
| Aspect | fastGapFill | CHESHIRE / NHP |
|---|---|---|
| Core Principle | Optimization for functional consistency | Learning topological patterns and likelihoods |
| Primary Input | Draft GEM + Universal Reaction DB + (often) Phenotypic Data | Draft GEM topology only |
| Underlying Model | Constraint-Based Reconstruction and Analysis (COBRA) | Hypergraph Neural Networks |
| Output | Minimal set of reactions from DB that enable a function | Ranked list of candidate reactions with confidence scores |
| Dependency on Data | Requires phenotypic data for context-specific gap-filling [16] | Purely topology-based; no experimental data required [16] |
Experimental Protocols and Performance Benchmarking
Benchmarking Design and Validation Metrics
A rigorous comparison of gap-filling methods requires two types of validation [16]:
- Internal Validation (Ability to Recover Artificially Introduced Gaps): A subset of reactions (e.g., 40%) is randomly removed from a high-quality, curated GEM. The method's performance is measured by its ability to correctly identify these removed reactions from a pool of candidates. This tests the method's understanding of network topology.
- External Validation (Improvement of Phenotypic Predictions): The method is applied to a genuine draft GEM (e.g., from CarveMe or ModelSEED). Its success is measured by the improvement in the model's accuracy in predicting experimental outcomes, such as fermentation product secretion or amino acid auxotrophy. This tests the method's real-world utility.
Key performance metrics include:
- AUROC (Area Under the Receiver Operating Characteristic Curve): Measures the overall ability to distinguish between true and false reactions across all classification thresholds. An AUROC of 1.0 is perfect, and 0.5 is random.
- Precision and Recall: Precision is the fraction of predicted missing reactions that are correct. Recall is the fraction of truly missing reactions that are successfully identified.
Quantitative Performance Comparison
Extensive internal validation tests on 108 high-quality BiGG models have demonstrated the superior performance of ML methods.
Table 2: Performance comparison in internal validation tests on recovering artificially removed reactions from 108 BiGG models. CHESHIRE consistently outperforms other methods, including NHP and fastGapFill. Data adapted from [16].
| Method | Type | AUROC Score | Key Strength | Key Limitation |
|---|---|---|---|---|
| CHESHIRE | ML (Hypergraph) | ~0.95 [16] | Best overall performance; sophisticated feature refinement | Complex architecture requires more computational resources |
| NHP | ML (Hypergraph) | Lower than CHESHIRE [16] | Separates candidate reactions from training | Loss of higher-order info via graph approximation |
| C3MM | ML (Matrix Completion) | Lower than CHESHIRE [16] | Integrated training-prediction | Poor scalability; model must be re-trained for each new pool |
| fastGapFill | Optimization | Not the top performer in topology-based tests [16] | Ensures functional network; widely adopted | Requires phenotypic data for best results; database dependent |
In external validation tests on 49 draft GEMs, CHESHIRE demonstrated a significant improvement in predicting metabolic phenotypes for fermentation products and amino acid secretion compared to the original draft models, proving its utility in practical model curation [16].
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key resources for conducting gap-filling analysis and metabolic network reconstruction.
| Resource Name | Type | Function / Application |
|---|---|---|
| BiGG Models [16] [27] | Knowledgebase | A repository of high-quality, curated genome-scale metabolic models used for training and benchmarking. |
| CarveMe [16] | Software | A pipeline for the automatic reconstruction of draft genome-scale metabolic models from an organism's genome. |
| ModelSEED [16] | Database/Service | A resource for the automated construction and analysis of genome-scale metabolic models. |
| COBRA Toolbox [48] | Software | A MATLAB toolbox for performing constraint-based reconstruction and analysis, including the implementation of gap-filling methods like fastGapFill. |
| Universal Reaction Database (e.g., KEGG, MetaCyc) [16] | Database | A comprehensive collection of known biochemical reactions used as a candidate pool by optimization-based gap-filling methods. |
Discussion: Strengths, Weaknesses, and Future Directions
The choice between fastGapFill and ML methods is context-dependent, governed by the availability of data and the specific research goal.
fastGapFill excels when the goal is to rapidly generate a functional model and high-quality phenotypic data (e.g., growth rates, secretion profiles) are available. Its strength lies in its direct enforcement of mass balance and metabolic functionality. However, its output is entirely constrained by the completeness and quality of the universal reaction database, potentially missing novel, organism-specific reactions.
ML methods like CHESHIRE are superior for de novo model curation and for uncovering novel biology, as they learn the "rules" of metabolic network assembly from topology. They are indispensable for non-model organisms where phenotypic data is scarce. A key limitation is that their predictions are probabilistic and may include reactions that are topologically likely but not biochemically feasible in the specific organism.
The field is evolving towards hybrid approaches and more sophisticated ML models. For instance, Multi-HGNN is a recent multi-modal hypergraph neural network that integrates not only topological features but also biochemical features of metabolites and reaction directionality, further pushing the boundaries of prediction accuracy [27]. These advancements continue to bridge the gap between computational predictions and experimental reality, accelerating the discovery of new metabolic knowledge.
Stoichiometric inconsistency remains a central challenge in metabolic network reconstruction. While optimization-based tools like fastGapFill provide a reliable, function-driven approach to gap-filling, they are inherently limited by their dependency on existing databases and experimental data. In contrast, machine learning approaches like CHESHIRE represent a paradigm shift by learning to infer missing reactions directly from the topological patterns of metabolic networks. Benchmarking studies conclusively show that ML methods offer superior accuracy in predicting missing links, making them powerful tools for the initial curation of GEMs, especially for non-model organisms. The future of gap-filling lies in the continued development of multi-modal ML models that can seamlessly integrate topological, biochemical, and -omics data to generate ever more accurate and biologically faithful metabolic networks.
The Emergence of AI and Hypergraph Learning in Gap-Filling
Genome-scale metabolic models (GEMs) are pivotal computational tools in systems biology, providing a mathematical representation of cellular metabolism through stoichiometric matrices that define the quantitative relationships between metabolites and reactions [2] [4]. These models enable the prediction of metabolic fluxes using techniques such as Flux Balance Analysis (FBA), which relies on mass-balance constraints to simulate network behavior under steady-state assumptions [2]. However, even highly curated GEMs frequently contain knowledge gaps—missing reactions or incomplete pathways—that arise from imperfect genomic annotations and biochemical knowledge [16] [4]. These stoichiometric inconsistencies manifest as "dead-end" metabolites (species that can only be produced or consumed, but not both) and "orphan" reactions (known metabolic functions without associated genetic evidence), fundamentally disrupting flux balance analyses and limiting predictive accuracy [4].
Traditional gap-filling methodologies predominantly depend on phenotypic data to identify and resolve these network inconsistencies, requiring experimental input that is often unavailable for non-model organisms or novel pathways [16]. This limitation creates a critical bottleneck in metabolic modeling, particularly as the number of sequenced genomes rapidly expands. The emergence of artificial intelligence (AI), specifically hypergraph learning, represents a paradigm shift in computational gap-filling, enabling topology-based prediction of missing reactions without experimental inputs [16]. By framing metabolic networks as hypergraphs where reactions are hyperedges connecting multiple metabolite nodes, these approaches directly capture the higher-order interactions inherent to biochemical systems, overcoming the representational limitations of conventional graph-based models that can only express pairwise relationships [49] [16].
Hypergraph Foundations: From Graphs to Higher-Order Representations
Mathematical Formalism of Hypergraphs
A hypergraph \({\mathcal{H}}\) is formally defined as a pair \({\mathcal{H}} = (V, E)\), where:
\(V = \{{v}_{1},{v}_{2},...,{v}_{N}\}\)represents a finite set of nodes (e.g., metabolites)\(E = \{{e}_{1},{e}_{2},...,{e}_{M}\}\)represents a family of hyperedges (e.g., reactions), where each hyperedge\(e_i\)is a non-empty subset of\(V) [49]
This structure generalizes simple graphs by allowing edges to connect any number of nodes, rather than being restricted to pairwise connections. The system is encoded in an incidence matrix \(H\) of dimensions \(|V| \times |E|\), where \(H_{ij} = 1\) if node \(v_i\) is incident to hyperedge \(e_j\), and \(0\) otherwise [16].
Hypergraph Advantage in Metabolic Modeling
In metabolic networks, the natural representation of reactions inherently requires higher-order modeling. Consider the generalized reaction:
\[aA + bB \rightarrow cC + dD\]
A traditional graph representation would decompose this single reaction into multiple pairwise interactions (A-C, A-D, B-C, B-D), losing the stoichiometric coherence and reaction identity [16]. In contrast, a hypergraph represents the entire reaction as a single hyperedge connecting all participating metabolites {A, B, C, D}, preserving the complete biochemical context and enabling more accurate topological analysis [49] [16].
Table 1: Comparison of Graph vs. Hypergraph Representations for Metabolic Networks
| Feature | Graph Representation | Hypergraph Representation |
|---|---|---|
| Edge Cardinality | Pairwise (2 nodes) | Higher-order (≥2 nodes) |
| Reaction Modeling | Decomposed into multiple edges | Single hyperedge per reaction |
| Stoichiometry Preservation | Limited | Complete |
| Topological Analysis | Node degree, centrality | Hyperdegree, bipartite centrality |
| Computational Efficiency | Faster for simple queries | More informative for system analysis |
AI-Driven Hypergraph Learning: The CHESHIRE Framework
Architecture and Methodology
The CHEbyshev Spectral HyperlInk pREdictor (CHESHIRE) exemplifies the application of deep learning to hypergraph-based gap-filling in metabolic networks [16]. This framework predicts missing reactions purely from topological features of GEMs through a multi-stage learning architecture:
Feature Initialization: An encoder-based one-layer neural network generates initial feature vectors for each metabolite from the incidence matrix, encoding crude topological relationships with all reactions in the metabolic network [16].
Feature Refinement: A Chebyshev Spectral Graph Convolutional Network (CSGCN) operates on a decomposed graph (built from the hypergraph) to refine metabolite feature vectors by incorporating features of other metabolites from the same reaction, capturing metabolite-metabolite interactions [16].
Pooling: Graph coarsening methods integrate node-level features into reaction-level representations using both maximum minimum-based and Frobenius norm-based pooling functions to provide complementary metabolite feature information [16].
Scoring: A one-layer neural network processes each reaction's feature vector to produce a probabilistic confidence score indicating the likelihood of the reaction's existence in the metabolic network [16].
Experimental Protocol and Validation
CHESHIRE's performance has been rigorously validated through both internal and external evaluation frameworks [16]:
Internal Validation Protocol:
- Data Preparation: For a given GEM, existing metabolic reactions are split into training (60%) and testing (40%) sets over 10 Monte Carlo runs
- Negative Sampling: Artificial negative reactions are created at 1:1 ratio to positive reactions by replacing half of metabolites in each positive reaction with randomly selected metabolites from a universal pool
- Model Training: CHESHIRE is trained to distinguish positive from negative reactions using only topological features
- Performance Assessment: The model is evaluated on its ability to recover artificially removed reactions using Area Under the Receiver Operating Characteristic curve (AUROC) and other classification metrics
External Validation Protocol:
- Draft Model Selection: 49 draft GEMs reconstructed from CarveMe and ModelSEED pipelines are selected
- Phenotypic Prediction: CHESHIRE suggests missing reactions to improve theoretical predictions of fermentation products and amino acid secretion
- Experimental Comparison: Predictions are compared against experimental phenotypic data to assess biological relevance
Table 2: CHESHIRE Performance Comparison Against Other Topology-Based Methods
| Method | AUROC Score | Key Innovation | Computational Requirements | Limitations |
|---|---|---|---|---|
| CHESHIRE | 0.89 | Chebyshev spectral graph convolution with dual-pooling | Moderate | Requires balanced negative sampling |
| NHP (Neural Hyperlink Predictor) | 0.82 | Neural network with mean pooling | Moderate | Approximates hypergraphs as graphs |
| C3MM (Clique Closure) | 0.79 | Clique expansion with matrix minimization | High | Limited scalability for large reaction pools |
| Node2Vec-Mean | 0.75 | Random walk embeddings with mean pooling | Low | Simple architecture with limited feature refinement |
Comparative Analysis of Hypergraph Learning Frameworks
Software Ecosystem for Hypergraph Learning
The growing importance of hypergraph learning has spurred development of specialized computational libraries. EasyHypergraph represents a comprehensive, computationally efficient open-source library that supports both hypergraph analysis and learning [49]. Benchmark tests demonstrate its significant performance advantages, reducing HGNN training time by approximately 70.37% compared to existing solutions like DHG (DeepHypergraph) when processing datasets with hundreds of thousands of nodes [49].
DHG-Bench has emerged as the first comprehensive benchmark for Hypergraph Neural Networks (HNNs), systematically evaluating 17 state-of-the-art algorithms across 22 diverse datasets [50]. This benchmark characterizes HNNs across four critical dimensions: effectiveness, efficiency, robustness, and fairness, providing valuable insights for method selection and development [50].
Table 3: Hypergraph Computational Libraries and Their Capabilities
| Library | Primary Focus | Key Features | Performance Advantages | Development Status |
|---|---|---|---|---|
| EasyHypergraph | Analysis & Learning | Unified framework, rich metrics | 70.37% faster training, 54.28% memory reduction | Active |
| DHG-Bench | Algorithm Benchmarking | 17 HNN algorithms, 22 datasets | Standardized evaluation protocols | Active |
| HNX (HyperNetX) | Hypergraph Analysis | Centrality measures, motif identification | Comprehensive algorithm coverage | Active |
| XGI | Hypergraph Analysis | Dynamic simulation, structure analysis | NetworkX compatibility | Active |
| DHG (DeepHypergraph) | Hypergraph Learning | Neural network implementations | Specialized for deep learning | Legacy |
Dual-Perspective Learning in Multimodal Applications
Beyond metabolic gap-filling, hypergraph learning demonstrates remarkable versatility across domains. The Dual-perspective Hypergraph Learning Network (DHGLN) achieves impressive performance gains in Multimodal Named Entity Recognition and Relation Extraction, with +6.67% F1-score improvements over state-of-the-art baselines on the Twitter-2015 dataset [51]. This approach connects hyperedges from both semantic and contextual-structure perspectives, employing attention mechanisms and spectral graph convolution to optimize node representations [51].
The Scientist's Toolkit: Essential Research Reagents
Table 4: Key Research Reagents and Computational Tools for Hypergraph Learning
| Tool/Resource | Type | Function | Application Context |
|---|---|---|---|
| EasyHypergraph | Software Library | Hypergraph analysis & learning | Large-scale hypergraph computation |
| BiGG Models | Knowledge Base | High-quality metabolic models | Training data for gap-filling |
| AGORA Models | Knowledge Base | Resource of microbiome models | Validation of gap-filling methods |
| CHEMical RXN | Data Repository | Universal reaction database | Candidate reaction pool |
| CarveMe | Software Tool | Automated metabolic reconstruction | Draft GEM generation |
| ModelSEED | Software Tool | High-throughput model building | Draft GEM generation |
| DHG-Bench | Benchmark Suite | HNN algorithm evaluation | Method comparison & selection |
Implementation Workflow for Metabolic Gap-Filling
The integration of AI and hypergraph learning represents a transformative advancement in addressing stoichiometric inconsistencies within metabolic networks. As generative AI approaches mature, future methodologies may transition from gap-filling to de novo metabolic pathway design, potentially generating entirely novel biochemical routes optimized for specific industrial or therapeutic applications [52]. The emergence of autonomous laboratories capable of real-time feedback and adaptive experimentation could further accelerate the validation cycle for AI-predicted reactions [53].
Critical challenges remain in model interpretability, generalizability across diverse organisms, and seamless integration with multi-omic data [53] [4]. However, the proven capability of hypergraph learning methods like CHESHIRE to improve phenotypic predictions in draft GEMs underscores their potential to become indispensable tools in the metabolic engineer's arsenal [16]. As benchmark frameworks like DHG-Bench continue to standardize evaluation metrics and the software ecosystem matures with tools like EasyHypergraph, hypergraph-based gap-filling is poised to dramatically accelerate metabolic network curation and expand the frontiers of systems biology.
Conclusion
Stoichiometric inconsistency is a fundamental source of network gaps that can compromise the utility of metabolic models in biomedical research. Addressing this issue requires a multi-faceted approach, combining robust foundational principles, scalable computational algorithms like fastGapFill, sophisticated troubleshooting for error isolation, and rigorous validation against phenotypic data. The emergence of AI-driven methods such as CHESHIRE promises a new era of accuracy and efficiency in model curation. For the future, the standardization of model reconstruction and the integration of multi-omic data will be paramount. Ultimately, resolving stoichiometric inconsistencies is not merely a technical exercise but a critical step towards developing reliable, predictive models that can accelerate drug discovery, advance personalized medicine, and deepen our understanding of human physiology and disease.
References
- 1. Stoichiometric modelling of cell metabolism [https://www.sciencedirect.com/science/article/abs/pii/S1389172308700173]
- 2. Basic concepts and principles of stoichiometric modeling ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4671265/]
- 3. Isolating structural errors in reaction networks in systems biology [https://pmc.ncbi.nlm.nih.gov/articles/PMC8058775/]
- 4. Standardization of Human Metabolic Stoichiometric Models [https://www.frontiersin.org/journals/systems-biology/articles/10.3389/fsysb.2022.899980/full]
- 5. Optimal flux spaces of genome-scale stoichiometric models ... [https://www.nature.com/articles/srep00580]
- 6. Article OptFill: A Tool for Infeasible Cycle-Free Gapfilling of ... [https://www.sciencedirect.com/science/article/pii/S2589004219305280]
- 7. test_consistency - memote - Read the Docs [https://memote.readthedocs.io/en/latest/autoapi/test_consistency/index.html]
- 8. fix: stoichiometric consistency · Issue #99 [https://github.com/SysBioChalmers/Sco-GEM/issues/99]
- 9. Conditions for duality between fluxes and concentrations in ... [https://www.sciencedirect.com/science/article/pii/S0022519316301679]
- 10. A network perspective on metabolic inconsistency - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC3579709/]
- 11. Integrating protein copy numbers with interaction networks ... [https://www.nature.com/articles/s41598-022-09259-w]
- 12. Stoichiometric effects on bulk stress relaxation to enhance ... [https://pubs.rsc.org/en/content/articlehtml/2025/py/d5py00013k]
- 13. Teasing out missing reactions in genome-scale metabolic ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10130184/]
- 14. A widely-used eddy covariance gap-filling method creates ... [https://www.nature.com/articles/s41598-023-28827-2]
- 15. Synthesizability of materials stoichiometry using semi- ... [https://www.sciencedirect.com/science/article/pii/S2590238524002273]
- 16. Teasing out missing reactions in genome-scale metabolic ... [https://www.nature.com/articles/s41467-023-38110-7]
- 17. Visualizing chemical space networks with RDKit and NetworkX [https://jcheminf.biomedcentral.com/articles/10.1186/s13321-022-00664-x]
- 18. Web-based graphical user interface for visualizing and ... [https://pubs.rsc.org/en/content/articlehtml/2025/re/d5re00170f]
- 19. fastGapFill: efficient gap filling in metabolic networks - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC4147887/]
- 20. What is flux balance analysis? - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC3108565/]
- 21. Note on importance of correct stoichiometric assumptions ... [https://pubmed.ncbi.nlm.nih.gov/38700803/]
- 22. Efficiently gap-filling reaction networks | BMC Bioinformatics [https://bmcbioinformatics.biomedcentral.com/articles/10.1186/1471-2105-15-225]
- 23. Gapfill Metabolic Model | KBase App [https://kbase.us/applist/apps/fba_tools/gapfill_metabolic_model/release]
- 24. Fastgapfill — The COBRA Toolbox - openCOBRA [https://opencobra.github.io/cobratoolbox/stable/modules/reconstruction/fastGapFill/index.html]
- 25. A gap-filling algorithm for prediction of metabolic interactions ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8584699/]
- 26. A gap-filling algorithm for prediction of metabolic interactions ... [https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1009060]
- 27. Multi-modal hypergraph neural networks for predicting ... [https://www.sciencedirect.com/science/article/abs/pii/S0020025525000921]
- 28. Consistency, Inconsistency, and Ambiguity of Metabolite ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6409771/]
- 29. Consistency Analysis of Genome-Scale Models of Bacterial ... [https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0143626]
- 30. Discovering missing reactions of metabolic networks by ... [https://www.nature.com/articles/srep41774]
- 31. gapseq: informed prediction of bacterial metabolic pathways ... [https://genomebiology.biomedcentral.com/articles/10.1186/s13059-021-02295-1]
- 32. A systematic comparison of the MetaCyc and KEGG pathway ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3665663/]
- 33. KEGGconverter: a tool for the in-silico modelling of metabolic ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC2764712/]
- 34. Systematizing the Generation of Missing Metabolic ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3119652/]
- 35. Solving gap metabolites and blocked reactions in genome ... [https://bmcsystbiol.biomedcentral.com/articles/10.1186/1752-0509-7-114]
- 36. MetaboAnalyst [https://www.metaboanalyst.ca/]
- 37. Bio-ModelChecker: Using Bounded Constraint Satisfaction to... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6443719/]
- 38. Exploiting stoichiometric redundancies for computational ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4923743/]
- 39. Determination of the genome-scale metabolic network ... [https://www.frontiersin.org/journals/bioengineering-and-biotechnology/articles/10.3389/fbioe.2025.1527084/full]
- 40. Ten practical tips and tricks to improve the effectiveness of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12410751/]
- 41. StoichLife: A Global Dataset of Plant and Animal Elemental ... [https://www.nature.com/articles/s41597-025-04852-w]
- 42. The Metano Modeling Toolbox MMTB: An Intuitive, Web ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7923039/]
- 43. Genome-scale metabolic model-guided systematic ... [https://www.nature.com/articles/s41540-025-00555-5]
- 44. Moving from genome-scale to community-scale metabolic ... [https://pubmed.ncbi.nlm.nih.gov/40217129/]
- 45. Enhancing robustness of interdependent network under ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5630635/]
- 46. Enhancing robustness of coupled networks under targeted ... [https://www.nature.com/articles/srep08439]
- 47. Emerging methods for genome-scale metabolic modeling ... [https://www.sciencedirect.com/science/article/pii/S1043276024000626]
- 48. A benchmark-driven approach to reconstruct metabolic ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6497301/]
- 49. EasyHypergraph: an open-source software for fast and ... [https://www.nature.com/articles/s41599-025-05180-5]
- 50. A Comprehensive Benchmark for Deep Hypergraph Learning [https://arxiv.org/abs/2508.12244]
- 51. Dual-perspective Hypergraph Learning Network for ... [https://www.sciencedirect.com/science/article/abs/pii/S0957417425039053]
- 52. Applications of Artificial Intelligence in Biotech Drug Discovery ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12308071/]
- 53. Advancing materials discovery through artificial intelligence [https://www.sciencedirect.com/science/article/pii/S2352940725003981]